Evaluation of a GADD45a-GFP human cell assay for rapid and reliable in vitro early genotoxicity screening
Posted: 9 April 2015 | Anne-Pascale Luzy (Galderma R&D), Guy Bouvier (Galderma R&D), Jean-Michel Linget (Galderma R&D), Nicolas Orsini (Galderma R&D)
This article evaluates a GADD45a-GFP human cell assay for rapid and reliable in vitro early genotoxicity screening…


As part of the drive to develop early screening strategies for improved drug discovery, a number of screening methods for the investigation of the genotoxic potential of molecules have been developed over the last few years. Unfortunately, few have proven to be sufficiently robust in practice and they do not all allow high throughput screening. As a result, only a limited number have undergone a programme of standardisation and validation using enough chemicals and sufficient participating laboratories to become of interest for industries or for regulatory purposes.
Genotoxicity is a concern in the pharmaceutical industry and there are a number of regulatory assays available to cover the different mechanisms of genotoxicity. These are deployed as a battery of in vitro and in vivo tests, as no single regulatory assay currently identifies all mechanisms of genotoxicity. Galderma, one of the fastest growing pharmaceutical companies within the EU, is focused exclusively within the field of dermatology and is committed to the research and development of new dermatological solutions. Improvements to genotoxicity screening strategy are an ongoing consideration at Galderma and by introducing a more specific and sensitive method for genotoxicity testing, this would enable genotoxicity screening to be carried out at an earlier stage of the drug development process than currently possible with traditional techniques. The GADD45a gene (Growth Arrest and DNA Damage gene) has been reported to have an important role in the maintenance of genomic integrity1. A new GADD45a-GFP human cell assay has been developed and has been proposed as a means of detecting genotoxic potential, based on the measurement of the Green Fluorescent Protein (GFP) expression under control of the of the GADD45a gene1,2. This study assessed the performance of this assay regarding its ability to optimise and minimise the number of screening tests required to determine genotoxicity in drug discovery screening.
The sensitivity and specificity of the GADD45a-GFP assay has been demonstrated in relation to the results of in vitro regulatory tests and to its prediction of rodent carcinogenesis1. Since then, several studies have investigated inter-laboratory reproducibility3,4, assessed assay performance in applicability domains for pharmaceutical compounds5, investigated metabolic activation6 and demonstrated its potential for high-throughput screening2,7,8 or optimisation of testing strategies to reduce testing with animals9.
Prior to our research, studies had been carried out comparing the assay with the ICH recommended in vitro genotoxicity tests – the bacterial Ames test, the mouse lymphoma assay (MLA) or chromosome aberration assay (CA) and the micronucleus assay (MNT). However, no information had ever been made available comparing the assay with early non-regulatory screening tests such as the SOS-ChromoTest, the Mini-Ames (using TA98 and TA100) and the in vitro screening micronucleus assay.
Only a limited number of studies have investigated the extent to which the GADD45a-GFP assay meets the specific needs of the pharmaceutical industry, i.e., the rapid optimisation of collections of drug candidates to eliminate compounds which are likely to have a genotoxic liability, to avoid the generation of misleading positive results in early screening and most importantly to avoid false negative results in later stage profiling. The study was undertaken to compare the GADD45a-GFP assay with the Mini-Ames, the SOS-ChromoTest and the in vitro screening MNT on a selection of proprietary compounds.
Our research was designed to help improve confidence and specificity in screening results, reduce the time taken for genotoxic profiling of drug candidates and decrease the amount of test material required. The primary aims were to enable Galderma to propose a refinement of its early genotoxicity screening battery and to establish the potential of the assay to clarify possible ambiguous results from other assays or to avoid misleading positive responses in relation to non-genotoxic mechanisms.
Regulatory requirements
Strict regulatory requirements for genotoxicity testing are in place for the pharmaceutical industry to ensure the safety of those administered with clinical candidates and approved marketed drugs. The potential for a drug substance to damage or alter DNA, thus causing genotoxicity and potentially cancer, is a significant area of toxicological concern. When assessing the regulatory safety of a pre-clinical drug candidate, it is essential that the testing regime is capable of accurately identifying genotoxic chemicals as well as detecting the different mechanisms by which genotoxicity occurs. Regulations therefore require testing both in vitro, in cell-based assays, and in vivo due to the fact that there is currently no single regulatory test capable of detecting all of the genotoxic mechanisms of action.
In addition, while the in vitro regulatory battery of tests provides a high level of detection of genotoxins and carcinogens, these tests have also been shown to generate many false or misleading positive results for non-carcinogens11. A high incidence of misleading positive results causes unnecessary delays to pre-clinical development and results in the need for additional testing to resolve data conflicts, which wastes time and money.
In 2011, the International Conference on Harmonisation (ICH) regulations, which cover testing of pharmaceuticals for human use, were amended to reflect changes to the existing battery tests with the aim of reducing false positives by lowering the top concentration for testing from 10mmol to 1mmol, and providing for an alternative option: e.g. in vitro Ames test followed by direct in vivo testing with measurement of two end points.
Early screening and the limitations of existing techniques
Regulatory genotoxicity testing has traditionally been carried out in the later stages of the drug discovery process. If issues are detected with the tested compounds, this can lead to high late-stage attrition, after the investment of significant R&D spend. As a result, companies are increasingly looking to adopt earlier testing for genotoxicity. The earlier testing methods were initially based on the regulatory battery of assays to provide sufficient coverage for the different modes of genotoxic action. As a consequence of moving the tests to an earlier stage of the discovery process, a higher number of compounds needed to be screened which has been achieved using both the Ames assay and various mammalian cell-based assays such as the MNT. While many organisations have implemented one of the screening Ames assays as a front line filter for genotoxicity, which provides an acceptable approach for the detection of mutagens, it is not comprehensive for all mechanisms and additional mammalian cell based testing is still required.
Ultimately, these screening assays still possess the limitations of regulatory assays, most notably with the unacceptable levels of false positives. Due to the lack of an assay strategy that provides high-throughput screening with the capability of identifying a broad range of genotoxic modes of action, coupled with a low false positive rate, conduct of comprehensive toxicological profiling earlier in the discovery phase represented a significant challenge. The early screening for genotoxicity is a key consideration for pharmaceutical lead molecule optimisation and the benefits are significant enabling the identification of development candidates more quickly and cost effectively.
There has been a lack of information available from companies regarding their in-house screening strategies but in recent years the GADD45a-GFP assay has been proposed as a rapid and accurate test for genotoxicity. This study involved the assessment of the assay to optimise and reduce the number of screening tests required to detect genotoxic hazard in early screening.
The performance of the assay was evaluated alongside the SOS-ChromoTest, the Mini-Ames (TA98 and TA100) and the screening micronucleus test to assess 20 selected proprietary compounds (A to T). For three of these (E, G and T), two or three different batches of the same compound were tested. The testing was carried out according to protocols as outlined in Hastwell1 and Jagger6 and all compounds were supplied coded with instructions for dilution if applicable. All compounds were prepared in an aqueous dilution comprising two per cent v/v dimethyl sulfoxide (DMSO).
The assay was performed on selected known molecules to test its predictability in relation to regulatory tests and to specifically investigate compounds with modes of action of interest to Galderma and confirm the potential of the assay to clarify in vitro genotoxicity profiles. The performance of the SOS-ChromoTest was not sufficient for the detection of genotoxic compounds especially compared with its low sensitivity for compounds detected as positive in the Mini-Ames, with a positive predictability value of 38%.
The full methods and results are presented in the Journal of Applied Toxicology paper entitled Evaluation of the GADD45α-GFP GreenScreen HC assay for rapid and reliable in vitro early genotoxicity screening (Table 1).
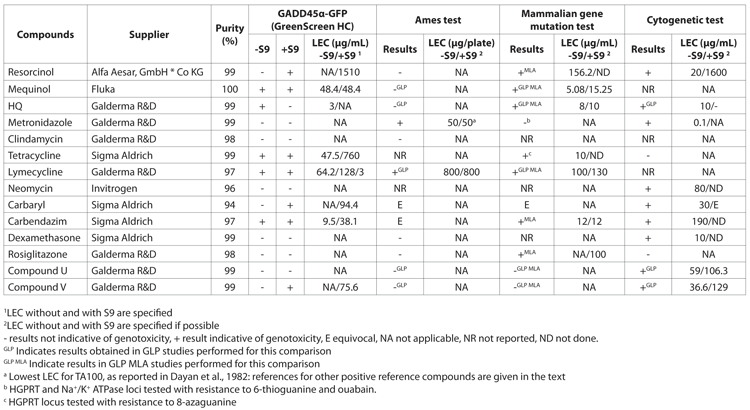

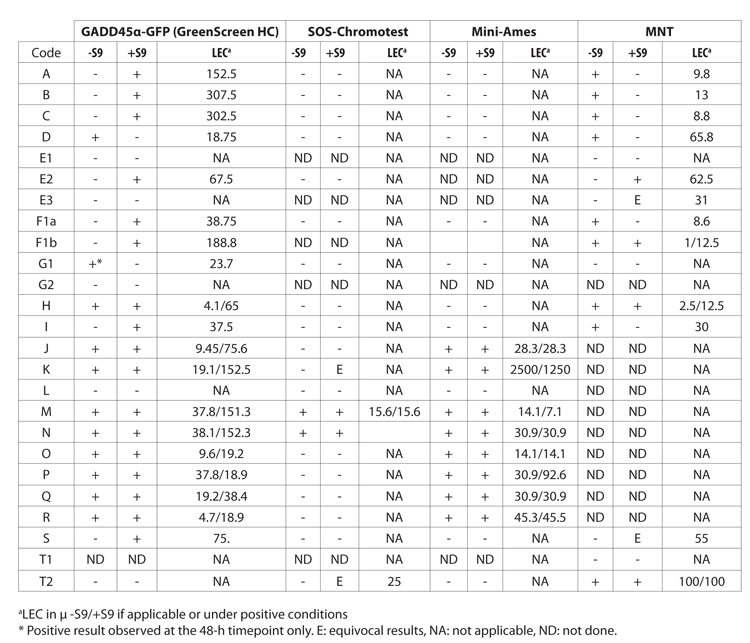

Table 1: Results of the assay, the SOS-Chomotest, the Mini-Ames and the micronucleus assay (MNT) for genotoxicity screening
Results and discussion: benefits of a GADD45a-GFP human cell assay
The introduction of reporter-based assays, such as the GADD45a-GFP assay, will help companies overcome the challenges of rapid and reliable in vitro genotoxicity screening. The assays were compared on the basis of their sensitivity, specificity, positive predictability, negative predictability and concordance; which are all key factors for companies looking to introduce early screening tests for genotoxicity.
The study concluded that the performance of the GADD45a-GFP assay as an early screening tool was superior to the SOS-ChromoTest in the detection of genotoxic compounds, although several GADD45a-GFP positive compounds in the assay were probably clastogens, and as such not necessarily expected SOS-ChromoTest positives. As the main purpose of this study was to evaluate the potential of the assay as a screening tool to accurately detect genotoxic compounds, this study compared the concordance of positive responses in the Ames test and the assay in other published articles. Although the screening data showed 100% sensitivity between positive Mini-Ames and assay results for proprietary compounds, results from the standard GADD45a-GFP assay reported in other studies suggest at least 75% (n = 12) sensitivity in general pharmaceutical space5, with the exception of the study by Olaharski10 in which reported a sensitivity of only 48%.
Among the ICH regulatory tests that were used in the comparative study, the MNT and the MLA were criticised for their high rate of false-positive results and poor specificity11. More recently, however, the combination of the Ames test with the in vitro micronucleus test was claimed sufficiently sensitive to detect rodent carcinogens12. Mammalian cell-based assays, such as the MNT and MLA, are sensitive to several confounding factors that may alter the relevance of the in vitro data for the in vivo evaluation as reviewed previously13. Thus, there was a particular interest for a screening strategy to investigate the potential of GADD45a-GFP to clarify results generated in mammalian tests that can be affected by some of these confounding factors.
The performance of the assay was assessed to clarify its effectiveness against confounding factors or ambiguous results from certain types of compounds, such as antibiotics, DNA synthesis inhibitors, aneugenic compounds, apoptosis inducers and the GADD45g inducer. Genotoxicity tests are often limited when it comes to the evaluation of antibiotics as their toxicity is sometimes difficult to evaluate in bacteria and the secondary pharmacology properties of antibiotics may lead to false-positive results. The results demonstrated the high specificity of the assay for GADD45a promoter and no interference with the GADD45g promoter and the effectiveness of the assay at clarifying confounding factors and ambiguous results.
The usefulness of the assay as a genotoxicity ranking tool and reproducibility of results was also assessed as optimising chemistry based on genotoxicity results can be easily performed if tests give repeatable results in terms of Lowest Effective Concentration (LECs) among and between replicates. Due to the fact that concentrations are based on cytotoxicity or solubility, such that not always the same testable concentrations are available for comparison, it is not always easy to compare LECs directly. The study carried out a comparison with a number of reference catecholamine molecules (hydroquinone (HQ), mequinol and resorcinol) as their genotoxic potential is supposed to be related to the length and position of the substitution groups on the phenol group and a decrease in the genotoxic potential was reported from HQ to resorcinol14. In agreement with previous studies, the GADD45a-GFP assay found the lowest LEC for HQ, followed by mequinol and lastly by resorcinol (Table 2). These results are also in agreement with the LECs obtained in mammalian gene mutation tests and cytogenetic tests, although results were not available for mequinol in all tests. The results confirmed the usefulness of GADD45a-GFP as a genotoxicity ranking tool with good reproducibility of results.
![Table 2: Comparison of results in the ‘GreenScreen’ HC (GS), the Ames test, the in vitro mammalian cell gene mutation assay and in vitro cytogenetic tests [chromosome aberration assay (CA) or micronucleus assay (MNT)] for selected compounds](https://www.drugtargetreview.com/wp-content/uploads/Evaluation-of-a-GADD45a-GFP-human-cell-assay-Table2.gif)
![Table 2: Comparison of results in the ‘GreenScreen’ HC (GS), the Ames test, the in vitro mammalian cell gene mutation assay and in vitro cytogenetic tests [chromosome aberration assay (CA) or micronucleus assay (MNT)] for selected compounds](https://www.drugtargetreview.com/wp-content/uploads/Evaluation-of-a-GADD45a-GFP-human-cell-assay-Table2.gif)
Table 2: Comparison of results in the ‘GreenScreen’ HC (GS), the Ames test, the in vitro mammalian cell gene mutation assay and in vitro cytogenetic tests [chromosome aberration assay (CA) or micronucleus assay (MNT)] for selected compounds
The assay was found to be highly effective in correctly identifying true positives and showed good concordance with other in vitro genotoxicity assays, giving pharmaceutical companies with extensive drug discovery operations a method for genotoxicity testing that can accommodate the demand for earlier positioning in the drug discovery process and the higher screening requirements of early profiling.
Conclusion
The GADD45a-GFP in this study (‘GreenScreen’ HC assay from Gentronix) demonstrated its usefulness as a screening tool in relation to other screening tests such as the SOS-ChromoTest, which was shown to be poorly efficient, the Mini-Ames, which gave parallel responses for all compounds tested, and the MNT, which gave comparable classification of compounds with regard to the genotoxic potential in all cases but one. We found the GADD45a-GFP assay to be a suitable and effective test for the early screening of drug candidates and considering the screening of reference molecules, provided the expected results and was able to clarify several conflicting results.
In this study, proprietary compounds F and V which were genotoxic in the assay were further evaluated non-genotoxic in vivo, leading to the conclusion that a positive GADD45a-GFP result is not always indicative of an in vivo genotoxic drug candidate. However, both compounds F and V also gave positive MNT/cytogenetic results in vitro and as such the GADD45A-GFP positive result was not unique. On the other hand, the assay testing conditions and results may be more representative of a true genotoxic profile.
Undetected toxicity can lead to late-stage project failure costing time and money. The GADD45a-GFP assay represents a solution to the problems surrounding the early identification of genotoxicity issues and their removal resulting in fewer toxic compounds being progressed and more effort being put towards compounds with a more robust safety profile. Ultimately this will lead to higher quality compounds being progressed through regulatory testing and into further investment. These new technologies will provide the pharmaceutical industry with new options and flexibility to address toxicity before they become a regulatory concern.
Further reading
For the full methods and results of the tests carried out please refer to the research article published in the Journal of Applied Toxicology entitled Evaluation of the GADD45α-GFP GreenScreen HC assay for rapid and reliable in vitro early genotoxicity screening – http://onlinelibrary.wiley.com/doi/10.1002/jat.2793/abstract
References
- Hastwell PW, Chai L-L, Roberts KJ, Webster TW, Harvey JS, Rees RW, Walmsley RM. 2006. High-specificity and high-sensitivity genotoxicity assessment in a human cell line: validation of the GreenScreen HC genotoxicity assay. Mutat. Res. 607: 160–175
- Knight AW, Birrel L, Walmsley RM. 2009a. Development and validation of a higher throughput screening approach to genotoxicity testing using the GADD45a-GFP GreenScreen HC assay. J. Biochem. Scr. 14: 16–30
- Billinton N, Hastwell PW, Beerens D, Birrell L, Ellis P, Maskell S, Webster TW, Windebank S, Woestenborghs F, Lynch AM, Scott AD, Tweats DJ, van Gompel J, Rees RW, Walmsley RM. 2008. Interlaboratory assessment of the GreenScreen HC GADD45a-GFP genotoxicity screening assay: An enabling study for independent validation as an alternative method. Mutat. Res. 653: 23–33
- Billinton N, Bruce S, Hansen, JR, Hastwell PW, Jagger C, McComb C, Klu, ML, Pant K, Rabinowitz A, Rees R, Tate M, Vinggaard AM, Walmsley RM. 2010. A pre-validation transferability study of the GreenScreen HC GADD45a-GFP assay with a metabolic activation system (S9). Mutat. Res. 700: 44–50
- Hastwell PW, Webster TW, Tate M, Billinton N, Lynch AM, Harvey JS, Rees RW, Walmsley RM. 2009. Analysis of 75 marketed pharmaceuticals using the GADD45a-GFP ‘GreenScreen HC’ genotoxicity assay. Mutagenesis 24: 455–463
- Jagger C, TateM, Cahill PA, Hughes C, Knight AW, Billinton N, Walmsley RM. 2009. Assessment of the genotoxicity of S9-generated metabolites using the GreenScreen HC GADD45a–GFP assay. Mutagenesis 24: 35–50
- Birrell L, Cahill P, Hughes C, Tate M, Walmsley RM. 2010. GADD45a-GFP GreenScreen HC assay results for the ECVAM recommended lists of genotoxic and non-genotoxic chemicals for assessment of new genotoxicity tests. Mutat. Res. 695: 87–95
- Knight AW, Little S, Houck K, Dix D, Judson R, Richard A, McCarrol N, Akerman G, Yang C, Birrel L, Walmsley RM. 2009b. Evaluation of high-throughput genotoxicity assays used in profiling the US EPA ToxCastTM chemicals. Regul. Toxicol. Pharma. 55: 188–199
- Lynch A, Sasaki JC, Elespuru R, Jacobson-Kram D, Thybaud V, De Boeck M, Aardema MJ, Aubrecht J, Benz RD, Dertinger S, Douglas GR, White PA, Escobar PA, Fornace A Jr, Honma M, Naven RT, Rusling JF, Schiest RH, Walmsley RM, Yamamura E, van Benthem J, Kim JH. 2011. New and emerging technologies for genetic toxicity testing. Environ. Mol. Mutagen. 52: 205–223
- Olaharski A, Albertini S, Kirchner S, Plats S, Uppal H, Lin H, Kolaja K. 2009. Evaluation of the GreenScreen GADD45a-GFP indicator assay with non-proprietary and proprietary compounds. Mutat. Res. 672: 10–16
- Kirkland D, Aardema M, Henderson L, Müller L. 2005. Evaluation of the ability of a battery of three in vitro genotoxicity tests to discriminate rodent carcinogens and non-carcinogens I. Sensitivity, specificity and relative predictivity. Mutat. Res. 584: 1–256
- Kirkland D, Reeve L, Gatehouse D, Vanparys P. 2011. A core in vitro genotoxicity battery comprising the Ames test plus the in vitro micronucleus test is sufficient to detect rodent carcinogens and in vivo genotoxins. Mutat. Res. 721: 27–73
- Scott D, Galloway SM, Marshall RR, Ishidate M Jr, Brusick D, Ashby J, Myhr BC. 1991. International Commission for protection against environmental mutagens and carcinogens. Genotoxicity under extreme culture conditions. A report from ICPEMC task group 9. Mutat. Res. 257: 147–204
- McGregor DB, Brown A, Cattanach P, Edwards I, McBride D, Caspary WJ. 1988. Response of the L5178Y tk+/tk- assay II: 18 coded chemicals. Env. Mol. Mutagen. 11: 91–118
Biographies
Anne-Pascale Luzy received her PhD in Human Biology from University of Chatenay Malabry Paris XI, about skin pharmacokinetic models to evaluate Pesticides exposure risk. She worked for 12 years at Aventis Crop Sciences in Sophia Antipolis Toxicology Center, before moving to Galderma R&D in 2002. She is now ADMET Screening and In Vitro Skin Penetration Unit Manager in early ADMET/early Formulation Group.
Jean-Michel Linget received a PhD in Pharmaco-Chemistry from Strasbourg University (1992). He reached Glaxo Research as DMPK Team leader, developing exploratory DMPK activities and supporting projects Elacridar and Tadalafil. He moved from GSK in 2003 for implementing DMPK at biotechs CareX then 7TM Pharma. Since 2009, he has been early ADMET/early Formulation Group Manager at Galderma. His main interests are the strategies for the selection of the best candidates and the optimal risk evaluation between Research and Development.
Guy Bouvier received his PhD in Human Biology from the University of Lyon, France, which he prepared conjointly between the team of Toxicology and Environmental Risk Factors of Professor Helmut Bartsch at the International Agency for Research on Cancer (Lyon France) and the Team of Professor Guy de Thé at the Faculty of Medicine. He worked as a scientist at the German Cancer Research Center (DKFZ) in Heidelberg before moving to industry since 1997, first with Rhone-Poulenc Agro. Since 2002, Guy has been working in product safety evaluation with Galderma.
Nicolas Orsini received a Biochemical University degree. He worked for three years in the Cellular and Molecular Pharmaco-toxicology Laboratory INSERM / INRA (UMR1064 ) and collaborated at two Europeans programmes, dealing with new environmental biomarkers, metabolism mechanisms, and toxicity related to water xenobiotics exposure. Since 2003, Nicolas has been working at Galderma in the early toxicology group and he is in charge of in vitro genotoxicity studies.
Related topics
Cell-based assays, Drug Discovery, Genomics, Screening
Related organisations
Galderma R&D
Related people
Anne-Pascale Luzy, Guy Bouvier, Jean-Michel Linget, Nicolas Orsini