The importance of characterising chemical starting points of drugs using appropriate in vitro ADME-toxicity assays
Posted: 21 March 2019 | Massimiliano Runfola - University of Pisa - Italy, Sheraz Gul - Fraunhofer IME - Germany | No comments yet
Over the past 30 years, one strategy the pharmaceutical industry has adopted in the drug discovery process has been to “fail early, fail often”.1,2 As most molecules in the early stages of drug discovery will have sub-optimal characteristics, significant modification is necessary to improve their properties.


For many reasons – including both scientific and business considerations – most of these compounds are not progressed. In order to mitigate risks early on, the practice of front-loading thousands of compounds and employing assays such as absorption, distribution, metabolism, excretion and toxicity (ADME-toxicity) at an early stage to select the most optimal compound for progression is often implemented. This strategy has been successful to some extent and here we illustrate the types of in vitro ADME-toxicity assays, which should be realised as early as possible in the drug discovery value chain.
The right ADME-toxicity assay
Billions of dollars are expended every year in the drug discovery process – especially for those pathologies characterised by inefficient or no available treatment, disease complexity, or unknown etiopathology, such as neurodegenerative diseases, metabolism-related disorders, cancer, and rare pathologies. However, among the many compounds that are selected as a lead, only a small proportion are successfully progressed to the candidate and clinical phases.2 As a consequence of the high attrition rates, the cost of developing a new drug is now reported to be more than $1 billion, taking over 10 years for a new treatment to be launched.3 A major cause of attrition is the sub-optimal toxicological and pharmacological characteristics of compounds, and it is estimated that around half of candidates fail in clinical trials due to unacceptable efficacy (largely ascribed to poor bioavailability due to ineffective intestinal absorption and undesirable metabolic stability) and toxicity.4 Therefore, identification of the pharmacokinetic and toxicological properties of compounds at an early stage using the appropriate assay systems would be expected to ensure the most optimal compounds are progressed to clinical trials, ultimately leading to a reduction in attrition. Many of the important in vitro ADME-toxicity assays that are employed to profile compounds in the early stages of drug discovery have relatively high throughput and these include cytotoxicity, cardiotoxicity, metabolic studies and off-target liabilities. This type of assay panel has been developed by the Fraunhofer IME, Germany and used in many projects to identify the most promising compounds for progression in the drug discovery value chain.5-15 These high-throughput compatible ADME-toxicity assays allow the profiling of hundreds of molecules in a relatively short time, thus allowing the selection of the most optimal compounds for structure activity relationship (SAR) studies in the hit-to-lead and lead optimisation process. These can be complemented with in silico predictions of pharmacological and toxicological properties and more recently through use of artificial intelligence methods to design compounds. We illustrate below the assays that should form part of an in vitro ADME-toxicity assay panel.
Cytotoxicity
One of the first steps in selecting a hit compound is the assessment of its cytotoxicity. Regarding in vitro models, a cytotoxic compound will often significantly interfere or alter cellular attachment, morphology, growth rate or induce cellular death.16 Many assays exist that can be used to monitor compound mediated toxicity. A commonly employed assay is to expose compounds to cells and measure parameters to provide an estimation of cell availability in correlation with the concentration of well-known metabolic factors, such as ATP or mitochondrial reductase potential, or with the activity of homeostatic housekeeping enzymes, eg, aminopeptidases. Cytotoxicity assays can also be formulated to detect loss of membrane integrity, a major marker of cellular death, or biomarkers closely related with cellular stress or with the early initiation of death mechanisms that are quantified by absorbance, luminescence or fluorescence spectrometry. Many of these biomarkers include proteases, which act upon specific fluorogenic or luminogenic peptide substrates, and lactate dehydrogenase (LDH).16
Cardiotoxicity
One of the reasons for drug failure in the clinic is due to adverse cardiac events. These are often difficult to identify, as their likelihood of occurrence is very low but can have life-threatening consequences. Several successful marketed drugs have either been given a black-box warning or withdrawn due to cardiotoxicity, especially related to QT-interval prolongation.17 The hERG channel plays a crucial role in regulating heart contractility and mediating the repolarising current in cardiac action potential; notably, guidelines have been recently published by the International Conference on Harmonization suggesting that all new drugs should be tested for hERG at the preclinical stage.17 One type of hERG assay makes use of the channels embedded in cell membrane preparations to which a fluorescent tracer molecule is bound and its displacement by a test compound is detected. Cardiotoxic compounds can inactivate hERG by displacing this tracer molecule, resulting in an increase in fluorescence polarisation, which can be easily measured. Moreover, novel assays exploiting embryonic stem cells or human induced pluripotent stem cell-derived cardiomyocytes have been developed with high-throughput capacity. These methods include calcium dynamics-based cardiomyocyte contractility assays and microelectrode array-based field potential assays.17
Metabolic studies
Beside toxicity, metabolism represents another important process that needs to be considered during pharmaceutical development, as it could explain drug-drug interactions, inter-subject variability, and toxic effects. Thus, several assays have been developed to assess metabolic stability for profiling of new chemical identities, as well as modulation of liver enzymes. One of the most important steps in the drug discovery process is the identification of enzymes involved in drug metabolism; particularly the cytochrome (CYP) superfamily, which represents the most important class of enzymes studied.
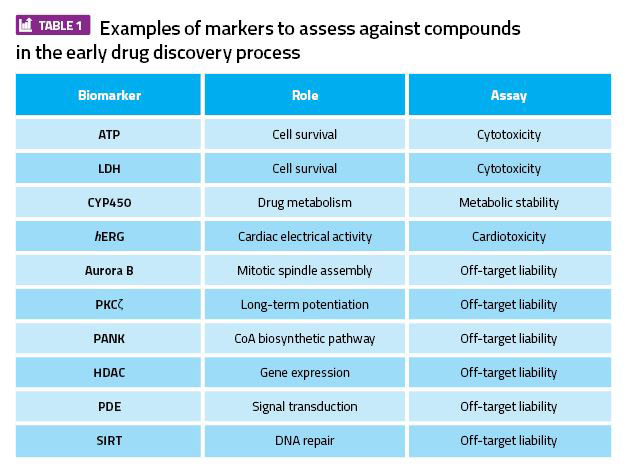

Table 1: Examples of markers to assess against compounds in the early drug discovery process.
Although more than 500 CYP isoforms have been identified in the human liver, only a few of them (CYP1A2, CYP2A6, CYP2C9, CYP2C19, CYP2D6, CYP2E1, and CYP3A4) are important for drug metabolism studies.4 For example, CYP3A4 is the most abundant CYP450 isoform and is involved in the metabolism of almost half of all marketed drugs as well as a high number of clinically significant drug-drug interactions.18 Screening compatible assays to measure the activities of CYP450 isoforms include luminescence and fluorescence techniques. The CYP enzymes are available as recombinant and native forms and can be used in cell-free inhibition assays as well as cell-based CYP450 induction assays. Different CYP biological matrices can be used for this purpose, including recombinant CYPs, liver microsomes, and hepatocytes. Among these, microsomes, which are subcellular fractions obtained by centrifugation of liver homogenates, are widely exploited to evaluate CYP inhibition and for drug stability studies.19 Due to their ease of handling, stability and availability (compared to other biological matrices), microsomes represent a model of choice for screening procedures, even if they are less accurate in providing information on in vivo metabolism than, for instance, hepatocytes.
Off-target liability
Another important step in identifying the optimal molecule for progression is the understanding of its ability to modulate well-known off-targets (or anti-targets). An off-target is defined as a biological target, which induces undesirable side effects when affected by a drug; for example, the hERG channel (described above) is a highly relevant off-target. Other important anti-targets include receptors for biogenic amines (eg, serotoninergic, adrenergic, and dopaminergic receptors), phosphodiesterases (PDEs), epigenetic regulators (HDACs and SIRTs), proteases and kinases. A candidate compound is often profiled against a large panel of anti-targets across the major drug target classes in order to obtain its fingerprint and likelihood of leading to adverse events in a clinical trial.1,2
Alternatively, a smaller panel of assays such as PDE, HDACs and specific kinases, especially those involved in cell cycle regulation (Aurora kinase, PANK, PKC, etc), can be used to profile compounds in order to reduce the costs and time when ranking compounds at an early stage (Table 1). An alternative to the profiling of a compound against individual targets is to use more sophisticated proteomic approaches – eg, kinome and genome profiling – which can provide a more phenotypically relevant picture of the liability of a compound.20
Conclusions
In conclusion, the profiling of a compound in a suitable in vitro ADME-toxicity assay panel is imperative when progressing compounds in the drug discovery value chain. These will allow the most optimal compounds with known pharmacokinetic and toxicological properties to be selected for progression to in vivo studies. Despite applying currently available methodologies, it often remains difficult to predict exactly how humans will respond when exposed to a potential drug. For this reason, novel approaches have recently been developed that use innovative model systems. For example, 3D cell cultures and organ-on-chip systems resemble organ functionalities more accurately than classical 2D cell cultures and their use could increase the number of optimal compounds entering clinical trials.21,22 Finally, novel systems exploiting zebrafish and nematode models could allow the identification of adverse events, thereby further ensuring that only the most optimal compounds are progressed in the drug discovery value chain.23,24
Biographies




References
1. Blomme EAG, Will Y. Toxicology Strategies for Drug Discovery: Present and Future. Chemical Research in Toxicology, 2016, 29, 473–504.
2. Waring MJ, et al. An analysis of the attrition of drug candidates from four major pharmaceutical companies. Nature Reviews Drug Discovery, 2015, 14, 475.
3. Tay-Teo K, et al. Comparison of Sales Income and Research and Development Costs for FDA-Approved Cancer Drugs Sold by Originator Drug Companies. JAMA Network Open, 2019, 2, e186875.
4. Li AP. Screening for human ADME/Tox drug properties in drug discovery. Drug Discovery Today, 2001, 6, 357–366.
5. Moraes CB, et al. Accelerating drug discovery efforts for trypanosomatidic infections using an integrated transnational academic drug discovery platform. SLAS Discovery, 2019, 24, 346–361.
6. Uliassi E, et al. Development of a focused library of triazole-linked privileged structure-based conjugates leading to the discovery of novel phenotypic hits against protozoan parasitic infections. ChemMedChem, 2018, 13, 678-683.
7. Lincianoa P, et al. Aryl thiosemicarbazones for the treatment of trypanosomatidic infections. European Journal of Medicinal Chemistry, 2018, 146, 423-434.
8. Spry C, et al. Structure-activity analysis of CJ-15,801 analogues that interact with Plasmodium falciparum pantothenate kinase and inhibit parasite proliferation. European Journal of Medicinal Chemistry, 2018, 1139-1147.
9. Uliassi E, et al. Design, synthesis and structure-activity relationships of a phenotypic small library against protozoan infections. MDPI Proceedings, 2017, 1, 648.
10. Uliassi E, et al. Crassiflorone derivatives that inhibit Trypanosoma brucei glyceraldehyde-3-phosphate dehydrogenase (TbGAPDH) and Trypanosoma cruzi trypanothione reductase (TcTR) and display activity against Trypanosoma brucei parasites. European Journal of Medicinal Chemistry, 2017, 141, 138-148.
11. Linciano P, et al. Exploiting the 2-amino-1,3,4-thiadiazole scaffold to inhibit Trypanosoma brucei pteridine reductase in support of early-stage drug discovery. ACS Omega, 2017, 2, 5666-5683.
12. von Delius M, et al. Synthesis of octaketides from the cytosporone family via C-H activation and investigation of biological activities. Israel Journal of Chemistry, 2017, 57, 975-981.
13. Di Pisa F, et al. Chroman-4-one derivatives targeting pteridine reductase 1 and showing anti-parasitic activity. Molecules, 2017, 22, E426.
14. Borsari C, et al. Methoxylated 2′-hydroxychalcones as antiparasitic hit compounds. European Journal of Medicinal Chemistry, 2017, 126, 1129-1135.
15. Borsari C, et al. Profiling of flavonol derivatives for the development of anti-trypanosomatidic drugs. Journal of Medicinal Chemistry, 2016, 59, 7598-7616.
16. Niles AL, et al. In vitro viability and cytotoxicity testing and same-well multi-parametric combinations for high throughput screening. Current chemical genomics, 2009. 3: p. 33-41.
17. Li X, et al. Cardiotoxicity screening: a review of rapid-throughput in vitro approaches. Archives of Toxicology, 2016, 90, 1803–1816.
18. Baranczewski P, et al. Introduction to in vitro estimation of metabolic stability and drug interactions of new chemical entities in drug discovery and development. Pharmacol Rep, 2006, 58, 453–72.
19. Lahoz A, et al. In vitro ADME medium/high-throughput screening in drug preclinical development. Mini reviews in medicinal chemistry, 2006, 6, 1053–1062.
20. Xiao Y, et al. A targeted quantitative proteomics strategy for global kinome profiling of cancer cells and tissues. Molecular & cellular proteomics: MCP, 2014, 13, 1065–1075.
21. Ishida S. Organs-on-a-chip: Current applications and consideration points for in vitro ADME-Tox studies. Drug Metab Pharmacokinet, 2018, 33, 49–54.
22. Nam KH, et al. Biomimetic 3D Tissue Models for Advanced High-Throughput Drug Screening.
J Lab Autom, 2015, 20, 201–15.
23. MacRae CA, Peterson RT. Zebrafish as tools for drug discovery. Nat Rev Drug Discov, 2015, 14, 721–31.
24. Perni M, et al. Massively parallel C. elegans tracking provides multi-dimensional fingerprints for phenotypic discovery. J Neurosci Methods, 2018, 306, 57–67.
The rest of this content is restricted - login or subscribe free to access


Why subscribe? Join our growing community of thousands of industry professionals and gain access to:
- quarterly issues in print and/or digital format
- case studies, whitepapers, webinars and industry-leading content
- breaking news and features
- our extensive online archive of thousands of articles and years of past issues
- ...And it's all free!
Click here to Subscribe today Login here
Related topics
Assays, Cell-based assays, Cytotoxicity assays, Drug Discovery, In Vivo
Related organisations
Fraunhofer-IME SP