Novel biomarker technology for cancer diagnostics
Posted: 11 September 2020 | Nikki Withers (Drug Target Review) | No comments yet
A new way of identifying cancer biomarkers has been developed by researchers at Lund University in Sweden. The new technology allows very sensitive, quick and cost-effective identification of cancer biomarkers. Nikki Withers spoke to Professor Carl Borrebaeck, who discusses how the researchers demonstrated the power of combining proteomics with genomics.

TODAY, EVERY third person will get cancer in their lifetime and the present trend suggests that in a few years that number will be one in two. If diagnosed earlier than is currently possible, a majority of cancer cases would have a much more favourable outcome for patients. The World Health Organization (WHO) has projected that a third of all cancers could be cured if diagnosed already at tumour stage I/II, that is, asymptomatic patients.
CREATE Health Cancer Center at Lund University has, in collaboration with Immunovia AB, developed a new technology combining the specificity of antibodies with the sensitivity of next-generation sequencing (NGS). The researchers believe the technology will pave the way for the next generation of biomarker discovery programmes in cancer, where there is still a tremendous unmet need.
“We have for years been developing advanced diagnostic approaches for multiplexed analysis of serum proteins, using a single drop of blood, for the purpose of early diagnosis of complex disease, in particular cancer,” said Professor Carl Borrebaeck, director of CREATE Health Cancer Center at Lund University. “There is a massive amount of information in blood that our combination of proteomics and genomics will open up for rapidly associating early tumour development with protein signatures. This in turn will benefit the patients with a more favourable outcome and overall survival.”
Challenges of analysing serum samples
Human serum is a complex proteome to analyse, creating major technological challenges for researchers. However, mining the serum proteome for differentially expressed molecular biomarkers provides an attractive and minimally invasive way for precision diagnostics. “Serum contains over 10,000 different proteins with a concentration range of 10e10 times, ie, a tremendous variety, which makes the analysis of crude unfractionated serum very difficult,” said Borrebaeck. However, the idea of removing a certain fraction of serum proteins before analysis is not recommended, since this would change the internal rations of diagnostically important serum proteins.
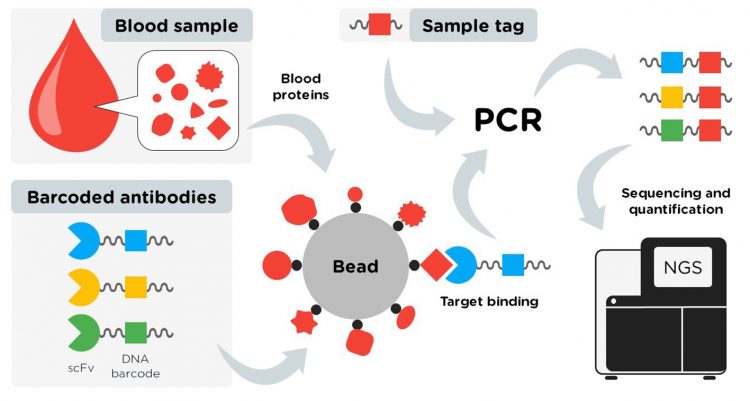

Figure 1: The concept of the ProMIS assay
The novel approach, denoted ProMIS, Protein detection using Multiplex Immunoassay in Solution, circumvents the inherent technical problems in conventional biomarker research traditionally utilising biomatrices, eg, planar- or bead-based arrays, by instead profiling serum proteins in solution. Since the entire process can be performed in solution, most inherent problems presented by using solid support are avoided.
ProMIS has a lot to contribute to cancer research and could actually show promise in the development or earlier and more accurate cancer diagnosis”
Borrebaeck explained that there are several technical advantages of profiling serum proteins in solution. Firstly, no solid/planar surfaces are used and the problem with unspecific adsorption is removed. It also does not require expensive, advanced spotting equipment. Researchers can also expect improved interactions between antibody and serum protein, resulting in an improved sensitivity due to less steric interferences. “Using NGS results in a dramatic increase in sensitivity. In theory, detection of one molecule is possible although not clinically relevant. However, it will allow us to dive deeper into the serum proteome,” he said.
Proof-of-concept
The research, which is published in Communications Biology,1 is a proof-of-concept study for profiling serum from pancreatic cancer patients, using ProMIS. The assay utilises antibody fragments (scFv) that are site specifically conjugated to DNA oligonucleotide barcodes, in a 1:1 manner, using a Sortase A-mediated coupling strategy. The barcoded scFvs are mixed with biotinylated serum proteins coupled to streptavidin-coated magnetic beads, and bound antibodies are detected, using NGS allowing for both a multiplex and sensitive read out.
The researchers noted that key to the concept is the oligonucleotide barcoding of the scFv antibodies that enables the sequencing readout. Discussing their findings, the team said: “The enzymatic reaction with Sortase A was found to be an effective and convenient strategy for conjugating scFv with a barcode in a 1:1 ratio under protein-compatible reaction conditions. Site‑specific conjugation avoids the risk of blocking the antigen binding site, which becomes a challenge in nonspecific conjugation methods. Multi-barcoding would limit the performance, which is also avoided with the 1:1 scFv:oligo conjugation principle of ProMIS.”
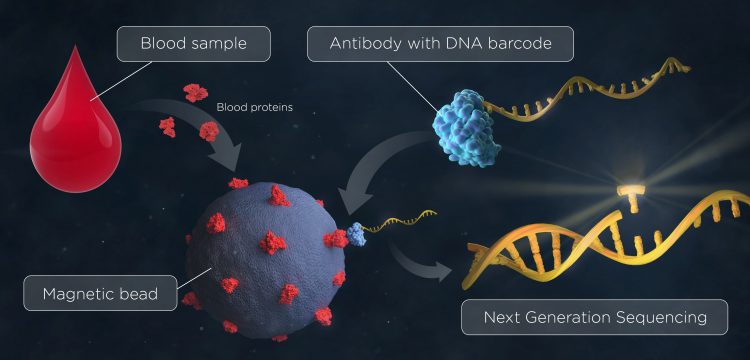

Figure 2: ProMIS circumvents the inherent technical problems in conventional biomarker research
The researchers concluded that the solution-based assay allows for easy conversion to a multi-well plate format that facilitates automation of the assay steps, which would provide consistent performance and a high sample throughput. In addition, with the PCR amplification step in combination with the NGS detection, it should be possible to tune the assay for excellent sensitivity.
Borrebaeck emphasised that cancer biomarker discovery and their association with different stages of disease is crucial for an improved therapeutic strategy and outcome for patients, such as overall survival. “With increased sensitivity, speed and lower cost of goods a completely new avenue of biomarker research will be available. Albeit in the beginning of development, ProMIS has a lot to contribute to cancer research and could actually show promise in the development or earlier and more accurate cancer diagnosis, which is a tremendous unmet clinical need.”
It is hoped that this novel approach, combining proteomics (antibodies) and genomics (NGS), will uniquely result in both a multiplex and ultra-sensitive read-out which in turn will increase the possibilities and success rate to find tumours earlier.
Acknowledgement
Images designed by study co-author, Mattias Brofelth


Reference
- Brofelth M, Ekstrand AI, Gour S. et al. Multiplex profiling of serum proteins in solution using barcoded antibody fragments and next generation sequencing. Commun Biol 3, 339 (2020).
Related topics
Assays, Biomarkers, Genomics, Next-Generation Sequencing (NGS), Oncology, Proteomics
Related conditions
Cancer
Related organisations
CREATE Health Cancer Center, Horizon, Immunovia AB, Lund University, World Health Organization (WHO)
Related people
Professor Carl Borrebaeck