Activity-directed synthesis: a structure-blind, function-driven molecular discovery approach
Posted: 16 December 2022 | Dr Niamh Morris (Rosalind Franklin Institute) | No comments yet
An autonomous strategy for molecular discovery could enable the exploration of a far broader chemical space than is possible with conventional approaches. In this article, Niamh Morris from Rosalind Franklin Institute outlines why activity-directed synthesis (ADS) could be the answer, providing examples of how this technique has the potential to enhance the hit discovery toolkit.

ADS is a function-driven approach to molecular discovery. Promiscuous reactions – with alternative possible products – are harnessed and the resulting crude product mixtures are directly screened for biological function. ADS thereby enables the parallel exploration of multiple molecular series and discovery of associated synthetic routes, making it an efficient way to screen against biological targets.
Traditional approaches to molecular discovery
Identifying bioactive molecules provides a starting point for the discovery of small molecule drugs to address a medical need or chemical probes with which to study biological mechanisms. However, searching chemical space for bioactivity is an enduring challenge that is exacerbated by its vastness (~1033 drug‑like molecules according to one estimate)1 and the poor coverage of the remarkably narrow reaction toolkit that currently underpins molecular discovery.2
In most molecular discovery projects, series of compounds are investigated one-by-one. These series are typically prepared using one of the reactions from the dominating reaction toolkit, restricting the chemical space that is explored. The toolkit has been expanded to include diversity-oriented methods3 which broaden the accessible chemical space. Roughly equal resources are invested in every molecule prepared, irrespective of whether they have the required biological function.
ADS: an alternative approach
ADS is a structure-blind, function-driven approach to molecular discovery (Figure 1). Promiscuous reactions are deliberately harnessed to generate mixtures of products. The crude reaction mixtures are directly screened for biological activity – an approach also taken by GSK in their direct-to-biology screening platform.4 Hit reactions that yield bioactive products are identified through screening. To optimise these hit reactions, the inputs (ie, substrates, solvent and catalyst) may be varied and the resulting reaction mixtures screened at progressively lower concentrations. This process can optimise the yield of the bioactive product or reveal more potent derivatives. The optimised reactions are ultimately scaled up and the products are purified, structurally elucidated and functionally characterised.
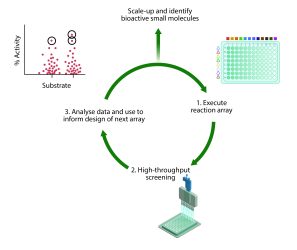

Figure 1: Workflow for ADS. Reaction arrays are performed to generate reaction mixtures that are screened crude for their biological activity. The data are analysed and reactions are prioritised on the basis of their biological performance alone. Following several rounds of ADS, optimised small molecules and associated synthetic routes are identified.
In ADS, resources are channelled into optimising reactions that have already proven to yield bioactive products, as opposed to every possible reaction and associated set of conditions. Following iterative cycles of ADS, a small molecule optimised for a given function and an associated synthetic route emerges.5 This function-driven emergence is analogous to the emergence of natural products via biosynthetic pathways,6 a concept that has also been harnessed in the synthetic fermentation strategy for identifying bioactive peptidomimetics.7 Like diversity-oriented methods, ADS enables broad exploration of chemical space by utilising promiscuous reactions outside of the standard medicinal chemistry toolkit. This allows for the discovery of multiple series in parallel as opposed to traditional one-by-one investigation of a molecular series.
ADS in action
ADS can facilitate a range of medicinal chemistry strategies. The approach was first applied in the discovery of novel androgen receptor (AR) modulators.6 In this work, metal-carbenoid chemistry was chosen due to the multiple possible outcomes enabled through both inter- and intramolecular reactions.
Table 1: ADS in action. Summary of case studies of ADS (adapted from (5)).
|
Medicinal chemistry application |
Chemistry |
Target (high-throughput assay) |
|
Scaffold discovery |
Metal carbenoid |
Androgen receptor (FRET)6 |
|
Fragment growth |
Metal carbenoid |
Androgen receptor (FRET)8 |
|
SAR study |
Pd-catalysed carbonylation |
Phenotypic antibacterial (S. aureus)9 |
|
Scaffold hopping |
Metal carbenoid |
p53/hDM2 (fluorescence anisotropy)11 |
Substrates were based on the 4-cyano-3-trifluoromethyl-phenyl motif, which is found in many known AR ligands. Reaction mixtures were subsequently screened in a FRET‑based high-throughput screen for AR agonists. Three rounds of ADS (involving 36, 192 and 108 reactions respectively) led to optimisation of synthetic yield and identification of three compounds with activity at sub-micromolar concentrations: two agonists (β-lactams 1 and 3) and one partial agonist (γ-lactam 2) (Figure 2). Crucially, neither of these chemotypes had previously annotated activity against AR.
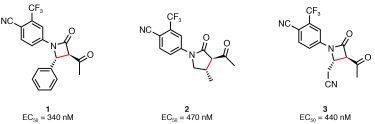

Figure 2: Novel AR modulators discovered using ADS. New bonds formed in the preparation of the ligands are shown in red.
The same chemistry and high-throughput screen were later used to drive the productive growth of an N-arylated acetamide fragment (Figure 3).8 In this case, reaction of the diazo substrates would enable growth of the fragment. These substrates were therefore reacted with diverse, commercially available co-substrates and the crude mixtures screened for biological function. Four distinct and novel chemotypes were identified with the most active product displaying a 125-fold increase in efficacy (12, EC50 = 730nM) compared to the original fragment 4. The diversity of these chemotypes stemmed from the many different reaction types possible with metal carbenoid chemistry.
More recently, the generality of ADS has been demonstrated using a phenotypic assay and a different reaction class. Here, Pd-catalysed carbonylation and an antibacterial screen (inhibition of S. aureus growth) were harnessed9 to expand a series of antibacterial quinazolinones.10 An exhaustive array of 220 reactions was executed and six of the resulting reaction mixtures exhibited substantial antibacterial activity (Figure 4).
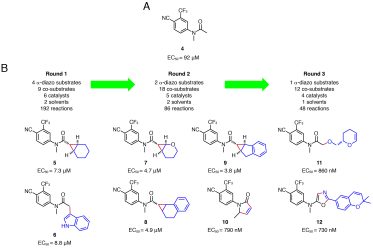

Figure 3: Activity-directed fragment-based discovery of AR ligands. The provenance of the ligands from diazo substrates (black) and co-substrates (blue) is shown, together with the new bonds (red).
Purification of the products of these reactions and structural elucidation revealed structure‑activity relationships for the series and compounds with MICs of between 0.016-2 µg/mL. Together, a range of case studies (Table 1) highlight the flexible nature of ADS in terms of its medicinal chemistry application, the underpinning chemistry and biological target.
Summary
ADS is a general, flexible approach to searching chemical space for bioactive small molecules, which simultaneously identifies an appropriate synthetic route. This generality, together with the focus on inherently bioactive compounds, makes ADS an attractive discovery approach. In the examples outlined above, all substrate design had the benefit of some prior knowledge of bioactivity, eg, using bioactive fragments as a basis for substrate design,6,8,11 or expanding a previously discovered antibacterial series.9 Future applications may harness larger target-agnostic arrays which could enable discovery of unprecedented chemotypes with unique modes of action.
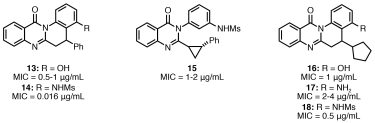

Figure 4: Activity-directed expansion of antibacterials. The MICs of six compounds active in the phenotypic screen against S. aureus strain ATCC
Additionally, the iterative nature of the process may enable autonomous ADS to be realised. Here, algorithms could be used to design all reaction arrays, with all experimental tasks executed robotically. A fully autonomous approach to molecular discovery would allow for a broader and even more diverse exploration of chemical space than is possible with conventional human-driven approaches – a feat that could open up new areas in biology by identifying novel targets and molecules with new mechanisms of action.
Niamh Morris
Niamh obtained her degree in Medicinal Chemistry from Trinity College Dublin in 2021. Niamh is a student on the Rosalind Franklin Institute PhD programme which is funded by UKRI-EPSRC. Her project focuses on the autonomous phenotype-directed discovery of chemical probes for the elucidation of biological mechanisms.
References:
- Polishchuk PG, Madzhidov TI, Varnek A. Estimation of the size of drug-like chemical space based on GDB-17 data. J Comput Aided Mol Des [Internet]. 2013 [cited 2022 Oct 26];27(8):675–9. Available from: https://pubmed.ncbi.nlm.nih.gov/23963658/
- Brown DG, Boström J. Analysis of Past and Present Synthetic Methodologies on Medicinal Chemistry: Where Have All the New Reactions Gone? J Med Chem [Internet]. 2016 May 26 [cited 2022 Oct 26];59(10):4443–58. Available from: https://pubs.acs.org/doi/full/10.1021/acs.jmedchem.5b01409
- Burke MD, Schreiber SL. A Planning Strategy for Diversity-Oriented Synthesis. Angewandte Chemie International Edition [Internet]. 2004 Jan 1 [cited 2022 Oct 26];43(1):46–58. Available from: https://onlinelibrary.wiley.com/doi/full/10.1002/anie.200300626
- Thomas RP, Heap RE, Zappacosta F, et al. A direct-to-biology high-throughput chemistry approach to reactive fragment screening. Chem Sci [Internet]. 2021 Sep 22 [cited 2022 Oct 26];12(36):12098–106. Available from: https://pubs.rsc.org/en/content/articlehtml/2021/sc/d1sc03551g
- Karageorgis G, Liver S, Nelson A. Activity-Directed Synthesis: A Flexible Approach for Lead Generation. ChemMedChem. 2020 Oct 5;15(19):1776–82.
- Karageorgis G, Warriner S, Nelson A. Efficient discovery of bioactive scaffolds by activity-directed synthesis. Nat Chem [Internet]. 2014 [cited 2022 Oct 12]; Available from: http://eprints.whiterose.ac.uk/80660/
- Huang YL, Bode JW. Synthetic fermentation of bioactive non-ribosomal peptides without organisms, enzymes or reagents. Nature Chemistry 2014 6:10 [Internet]. 2014 Sep 7 [cited 2022 Oct 26];6(10):877–84. Available from: https://www.nature.com/articles/nchem.2048
- Karageorgis G, Dow M, Aimon A, et al. Activity-Directed Synthesis with Intermolecular Reactions: Development of a Fragment into a Range of Androgen Receptor Agonists. Angewandte Chemie – International Edition. 2015 Nov 1;54(46):13538–44.
- Leggott A, Clarke JE, Chow S, et al. Activity-directed expansion of a series of antibacterial agents. Chemical Communications [Internet]. 2020 Jul 21 [cited 2022 Oct 12];56(58):8047–50. Available from: https://pubs.rsc.org/en/content/articlehtml/2020/cc/d0cc02361b
- Bouley R, Ding D, Peng Z, et al. Structure-Activity Relationship for the 4(3H)-Quinazolinone Antibacterials. J Med Chem [Internet]. 2016 May 26 [cited 2022 Nov 1];59(10):5011–21. Available from: https://pubs.acs.org/doi/full/10.1021/acs.jmedchem.6b00372
- Green AI, Hobor F, Tinworth CP, et al. Activity-Directed Synthesis of Inhibitors of the p53/hDM2 Protein–Protein Interaction. Chemistry – A European Journal. 2020 Aug 21;26(47):10682–9.
Related topics
Drug Discovery, Drug Discovery Processes, Molecular Biology, Molecular Targets, Technology
Related organisations
Trinity College Dublin