Unlocking a new molecular space in rapid drug discovery
Posted: 23 October 2024 | Dr Jorg Scheuermann (ETH Zurich), Dr Michelle Keller (ETH Zurich) | No comments yet
So-called undruggable pharmaceutical targets could be tackled with intermediate sized drugs such as macrocycles, combining the advantages of small molecules and biologics. The preparation of very large libraries of chemically synthesised intermediate-sized molecules is now in reach with a new technology developed at ETH Zurich. In this article, Dr Michelle Keller and Dr Jorg Scheuermann explain more.

Technologies for the rapid and efficient testing of small molecules and biologics have greatly accelerated drug discovery. However, many pharmaceutically relelvant targets remain ‘undruggable’. In particular, targeting intracellular protein-protein interactions is a challenge, as biologics have difficulty entering cells and small molecules often do not exhibit sufficient binding affinity and specificity to extended protein surfaces. Intermediate-sized molecules such as macrocycles combining the beneficial properties of both small molecules and biologics may enable the targeting of currently undruggable targets. However, until now, the technology allowing for the rapid, efficient, and cost-effective testing of millions to billions of intermediate‑sized chemically synthesised complex molecules did not exist.
Limitations of DNA-encoded chemical libraries in drug discovery
DNA-encoded chemical libraries (DEL) are widely used in the pharmaceutical industry for drug discovery of small molecules.1, 2 They allow the screening of millions of molecules for affinity to a target in a single experiment by encoding each small molecule with a unique DNA barcode.3 Molecules binding to a pharmaceutically relevant target can be fished out of a library pool of millions of molecules, providing a cheap and efficient means to discover new ligands. While DEL technology has provided valuable tool compounds and drug candidates, it is still limited in terms of the types of molecular structures that can be tested. DEL are synthesized by split-and-pool methodology, usually combining a maximum of 2–4 building block sets to assemble the displayed full molecules, recording each synthetic step by enzymatic ligation of DNA barcodes. With increasing molecule complexity, achieved by a greater number of synthesis steps, the yield of the final desired molecules decreases while truncated side‑products arise. This causes DEL to be mixtures of desired molecules and impurities, hampering hit identification. To date, there is no analytical means to assess the display quality of a DEL. This leads to unreliable selection results with poor signal-to-noise ratios in affinity-based selections for complex DEL, thereby limiting DEL technology in molecule diversity and managable library sizes such that it is not suited for testing intermediate-sized molecules.
The Pure-DEL Synthesis Platform
The DEL technology group at ETH Zurich has developed a new “Pure-DEL” synthesis platform providing inherently pure libraries, as recently described in Science.4 Thanks to self-purification of molecules after the last synthesis step, only library members with correctly assembled molecules are included in the final library (Figure 1). While conventional DEL synthesis is performed in solution, Pure-DEL synthesis is performed on solid support in combination with a dual-linker purification strategy. Split-and-pool synthesis is performed to assemble the desired molecules, capping unreacted starting material after every reaction. Following synthesis, only the properly assembled molecules which are not capped are connected to the solid support through a second orthogonally cleavable linker. By cleaving the first tether, all truncates are removed while only the pure library remains on the solid support and can subsequently be collected by cleaving the second linker and used in affinity-based selections against targets of interest.
Biomarkers are redefining how precision therapies are discovered, validated and delivered.
This exclusive expert-led report reveals how leading teams are using biomarker science to drive faster insights, cleaner data and more targeted treatments – from discovery to diagnostics.
Inside the report:
- How leading organisations are reshaping strategy with biomarker-led approaches
- Better tools for real-time decision-making – turning complex data into faster insights
- Global standardisation and assay sensitivity – what it takes to scale across networks
Discover how biomarker science is addressing the biggest hurdles in drug discovery, translational research and precision medicine – access your free copy today
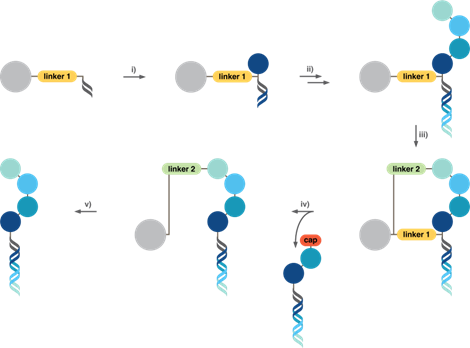

Figure 1: Synthesis process of self-purified DNA-encoded chemical libraries by dual-linker solid phase synthesis. Synthesis commences with library members connected to the solid support (large grey circle) through a first cleavable linker. i) A first cycle of split-and-pool synthesis is performed, splitting the material into separate compartments for incorporating a first set of chemical building blocks (dark blue circle), capping unreacted starting material, encoding each building block by enzymatic ligation of DNA barcodes, and then pooling all material together. ii) Further cycles of split-and-pool synthesis are performed until the desired molecular structure is obtained. iii) After synthesis is completed, the last buildling block is connected to the solid support through a second cleavable linker. Only those library members which have been properly assembled are connected to the solid support through two tethers, as incompletely synthesised capped truncated structures cannot react with the second linker. iv) The two linkers are orthogonally cleavable, such that by cleaving the first linker, all truncates are removed from the solid support, while only the pure library members remain attached to the solid support through the second linker. v) After washing away the capped impurities, the final purified library is collected by cleaving the second linker.
Dual-linker solid phase DEL synthesis provides, for the first time, a direct correlation of chemical display and encoding in DEL technology. While for conventional DEL, DNA barcodes are present in the final library regardless of whether the correct molecule is displayed or not, Pure-DEL libraries are only made up of library members with fully synthesised molecules displayed. By sequencing the DNA barcodes of a Pure-DEL, a clear picture of every molecule present in the library is obtained. In affinity-based selections against a target, reliable selection data can be attained, with hits clearly distinguished from the background. This technology also promises meaningful selection results for libraries of much larger sizes than currently possible – in the range of billions – and the display of complex intermediate-sized molecular structures with a high chance at oral bioavailability, proteolytic stability, cell permeability and binding to ‘undruggable’ targets.
Unlocking new possibilities
An additional advantage of self-purified DEL is that chemical reactions on the solid support can be performed in water-free conditions, in contrast to conventional DEL technology which requires aqueous conditions for solubilisation of the DNA barcode. This opens up the use of a much broader range of reaction types, allowing for the synthesis of highly diverse molecules spanning an underexplored chemical space. Pure-DEL libraries are not restricted to the incorporation of mainly natural amino acids, as in display technologies based on the natural translation machinery but can in principle be assembled by using any reaction or building block type. Libraries of intermediate-sized molecules with drug-like properties focusing on cell permeability and potential oral availability are now in reach, promising to finally tackle ‘undruggable’ targets.
References
1 Satz AL, Brunschweiger A, Flanagan ME, Gloger A, Hansen NJV, Kuai L, et al. DNA-encoded chemical libraries. Nature Reviews Methods Primers. 2022 Jan 17;2(1).
2 Keller M, Schira K, Scheuermann J. Impact of DNA-Encoded Chemical Library Technology on Drug Discovery. CHIMIA. 2022 May 25;76(5):388.
3 Neri D, Lerner RA. DNA-Encoded Chemical Libraries: A Selection System Based on Endowing Organic Compounds with Amplifiable Information. Annual Review of Biochemistry. 2018 Jun 20;87(1):479–502.
4 Keller M, Petrov D, Gloger A, Bastien Dietschi, Jobin K, Timon Gradinger, et al. Highly pure DNA-encoded chemical libraries by dual-linker solid-phase synthesis. Science. 2024 Jun 14;384(6701):1259–65.
About the authors
Dr Michelle Keller, ETH Zurich




PD Dr Jörg Scheuermann studied Chemistry at the University of Heidelberg, Germany and at ETH Zurich, Switzerland. Following his PhD studies at the ETH under the supervision of Prof. Dario Neri, he co-pioneered the development of DEL technology in different implementations together with Professor Dario Neri. He has published over 65 peer-reviewed manuscripts on DEL and is co‑founder and organiser of the International Symposium on DNA-Encoded Chemical Libraries. PD Dr Scheuermann is Principal Investigator at ETH Zurich, heading a group of four PhD students, two Postdocs and one Senior Scientist.
Related topics
Biologics, Drug Discovery, Drug Discovery Processes
Related organisations
ETH Zurich
Related people
Dr Jorg Scheuermann (ETH Zurich), Dr Michelle Keller (ETH Zurich)