Scientific workflow for hypothesis testing in drug discovery: part 3 of 3
Posted: 14 February 2025 | Dr Raminderpal Singh (Hitchhikers AI and 20/15 Visioneers), Nina Truter | No comments yet
Drug discovery scientists develop and test complex hypotheses using data and expertise, and build workflows to support this. In this third and final article, Dr Raminderpal Singh and Nina Truter summarise the tools used in the scientific workflow – and include key considerations.


Throughout the workflow described in Figure 1, different tools can play a critical role in facilitating each stage of analysis. From hypothesis generation to data cleaning and interpretation, the appropriate use of tools can significantly improve the efficiency and accuracy of the research process.
1. Data visualisation and hypothesis generation tools
Tools like Miro, that generate diagrams, are essential for mapping out hypotheses. Miro allows researchers to create a visual representation of the relationships between proteins, genes or pathways, helping to clarify the expected interactions within the biological system being studied. This kind of image is particularly useful during the hypothesis generation phase, where researchers are still exploring the relationships between different biological components.
ChatGPT is useful for brainstorming and generating new research ideas, and can also be used to explore possible pathways or protein interactions by inputting key terms or genes. This tool, while useful for generating ideas, should be used cautiously. Though it can provide new pathways or hypotheses to investigate, it should not replace rigorous literature review or empirical evidence.
2. Data cleaning and descriptive analytics tools
Excel remains one of the most commonly used tools for data cleaning and descriptive analytics in many research settings. Researchers use Excel for tasks such as sorting data, identifying outliers, and generating basic plots. However, for larger datasets, Excel has its limitations in terms of both scalability and complexity. Tools like R and Python, with libraries such as Pandas for data manipulation and Matplotlib for visualisation, provide robust solutions for handling large datasets and performing advanced statistical analyses. Python’s SciPy and Statsmodels libraries, for example, offer advanced tools for hypothesis testing, regression analysis and other complex statistical procedures that surpass Excel’s capabilities. ChatGPT and Claude.ai are useful tools to empower scientists with no coding experience by providing custom-written code for specific analyses and execution of this code. Again, this is not a replacement for rigorous analyses by data scientists; however, where data scientists are not available, it enables exploration of the data beyond the capabilities of Excel.
Another powerful workflow tool is the KEGG Pathway database, which helps researchers map out how proteins and genes interact within known biological pathways. This is especially useful during the hypothesis testing phase, as it enables researchers to visualise how their proteins of interest fit into broader biological processes. The KEGG Pathway database provides insights into metabolic pathways, genetic interactions and disease mechanisms, which are crucial for understanding how a dataset can inform our understanding of complex biological phenomena such as signal transduction, cell proliferation, or immune responses.
Gene ontology databases, such as STRING and Reactome, are additional tools that can be used to understand protein-protein interactions and their involvement in cellular processes. These tools are essential for interpreting the results of data analysis, particularly when the dataset reveals unexpected or novel interactions between proteins that require further investigation.
3. Network and interaction mapping tools
As biological datasets grow in complexity, graph-based tools have become essential for visualising and analysing protein-protein interactions and gene networks. Cytoscape, for example, is a widely used software tool for visualising molecular interaction networks and integrating these with gene expression profiles and other data. In research focused on drug discovery, understanding the interactions between multiple proteins or genes are crucial for identifying potential drug targets or understanding the mechanisms behind drug resistance.
Network-based approaches are also becoming more prevalent as researchers aim to represent complex biological data in more intuitive ways. By visualising data as networks or graphs, scientists can more easily identify hubs, bottlenecks or key players in biological processes, allowing them to focus their efforts on the most critical components of a system.
4. Literature and data curation tools
Data curation is a key part of any workflow, particularly when working with large datasets or integrating data from multiple sources. Tools like GeneCards are useful for obtaining detailed information about genes and their functions. GeneCards offers comprehensive gene-related information, such as pathways, interactions and diseases associated with each gene. This information is invaluable when generating hypotheses or validating findings, since it provides a deeper understanding of how a particular gene or protein fits into the broader biological context.
In addition to GeneCards, tools like Mendeley and Zotero are beneficial for managing research papers and references, particularly for researchers who rely on literature reviews to support their hypotheses and analyses. Proper reference management ensures efficient source tracking and maintains the integrity of work.
5. AI and machine learning tools
As biological research datasets grow in size and complexity, the use of AI and machine learning tools becomes more critical. ChatGPT can function as a brainstorming tool for generating hypotheses or exploring possible pathways and while this tool is still relatively novel in the research community, it represents the growing intersection between AI and drug discovery. ChatGPT can assist by summarising literature, suggesting new angles of inquiry, or even helping to explore large datasets in ways that would be too time-consuming for manual review.
Other machine learning tools, such as TensorFlow or PyTorch, can be used to analyse large datasets and identify patterns that may not be immediately apparent through traditional methods. These tools allow researchers to build predictive models, classify data, or identify novel associations between variables. In drug discovery, machine learning models have been used to predict drug efficacy, optimise compound structures and even simulate biological systems.
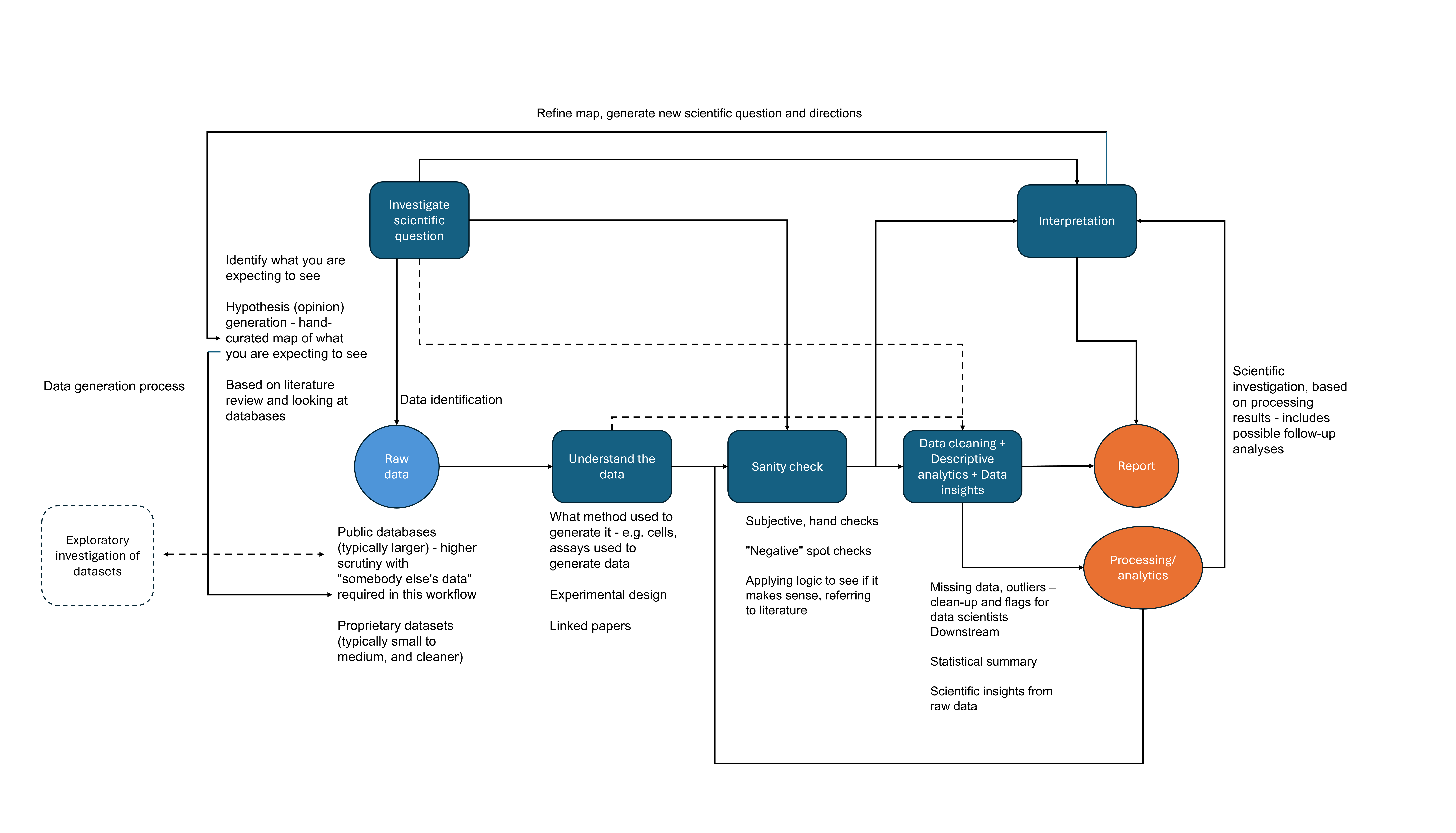

Figure 1: An illustration to demonstrate a high-level workflow for early drug discovery.
Summary of tools and databases used in the workflow:
- KEGG Pathway database – The KEGG (Kyoto Encyclopedia of Genes and Genomes) Pathway database provides information on molecular interaction and reaction networks for various biological pathways. https://www.kegg.jp/kegg/pathway.html
- STRING database – a database of known and predicted protein-protein interactions, integrating both physical and functional associations. https://string-db.org
- Reactome – an open-source, curated pathway database that provides insights into biological processes and molecular interactions. https://reactome.org
- GeneCards – a comprehensive database that provides detailed information on all known and predicted human genes, including functions, pathways and related diseases. https://www.genecards.org
- Cytoscape – a software platform for visualising molecular interaction networks and integrating these networks with gene expression profiles and other data. https://cytoscape.org
- Mendeley – a reference manager and academic social network that helps researchers organise research papers, collaborate online and discover the latest scientific research. https://www.mendeley.com
- Zotero – a free, easy-to-use tool to help researchers collect, organise, cite and share research. https://www.zotero.org
- TensorFlow – an open-source platform for machine learning, commonly used for deep learning applications and large dataset analysis. https://www.tensorflow.org
- PyTorch – an open-source machine learning library based on the Torch library, used for applications such as computer vision and natural language processing. https://pytorch.org
About the author
Dr Raminderpal Singh


Dr Raminderpal Singh is a recognised visionary in the implementation of AI across technology and science-focused industries. He has over 30 years of global experience leading and advising teams, helping early to mid-stage companies achieve breakthroughs through the effective use f computational modelling. Raminderpal is currently the Global Head of AI and GenAI Practice at 20/15 Visioneers. He also founded and leads the HitchhikersAI.org open-source community. He is also a co-founder of Incubate Bio – a techbio that helps life sciences companies accelerate their research and lower their wet lab costs through in silico modelling.
Raminderpal has extensive experience building businesses in both Europe and the US. As a business executive at IBM Research in New York, Dr Singh led the go-to-market for IBM Watson Genomics Analytics. He was also Vice President and Head of the Microbiome Division at Eagle Genomics Ltd in Cambridge. Raminderpal earned his PhD in semiconductor modelling in 1997. He has published several papers and two books and has twelve issued patents. In 2003, he was selected by EE Times as one of the top 13 most influential people in the semiconductor industry.
For more: http://raminderpalsingh.com; http://20visioneers15.com; http://hitchhikersAI.org; http://incubate.bio
Nina Truter
Nina Truter is a translational scientist with a deep focus on understanding mechanisms of action in drug development and leveraging disparate datasets in biotech. Based in South Africa, she has worked extensively with international biotech companies, specialising in therapeutic development for ageing-related diseases and complex conditions such as glioblastoma and Autosomal dominant polycystic kidney disease (ADPKD).
Her recent work includes consulting for UK-based biotech firms and leading initiatives in HitchhikersAI.org to advance the translation of AI and data science into practical biotech solutions such as identifying combination therapy opportunities and enhancing patient selection. In her work, she uses a systems approach to integrate insights from diverse datasets across in vitro, in vivo and human models to answer critical scientific questions, and translates biological mechanisms into models that are used by advanced analytical methods such as Pearlian causal inference.
For more: https://njtruter.wixsite.com/ninatruter
Related topics
Bioinformatics, Drug Discovery, Drug Discovery Processes, Machine learning
Related people
Dr Raminderpal Singh, Nina Truter