A high-throughput approach for measuring intracellular ATP levels to access compound-mediated cellular toxicity
Posted: 30 November 2018 | Maria Kuzikov (Fraunhofer-IME SP), Oliver Keminer (Fraunhofer Institute for Molecular Biology and Applied Ecology), Salma Hachim (Fraunhofer Institute for Molecular Biology and Applied Ecology), Sheraz Gul (Fraunhofer-IME SP) | No comments yet
Many drugs fail in clinical trials due to toxicity-related issues; therefore, drug-mediated cellular toxicity should be determined at an early stage in the drug discovery value chain. There are many assay systems that can be utilised to measure cellular toxicity. In this article we demonstrate that intracellular ATP concentrations are a sensitive method with which to determine cellular toxicity and, as a validation exercise, we report a screening compatible cellular toxicity assay protocol. More importantly, we report the output of a Proof-of-Concept screen that made use of this assay against a known drug library and the MCF-7 cell-line. The results will be beneficial for those engaged in drug repurposing.
An understanding of the ADME-tox profile of a compound is important when deciding if it should be progressed in the drug discovery value chain. Failure to identify potential problems with a compound at an early stage will make it more likely to fail in the clinic. In order to mitigate such risks, cellular assay systems are being extensively used in screening campaigns against diverse compound libraries to identify chemical starting points for drugs.1,2 The hits from these screens are generally weakly active with IC50in the low micromolar range and a major issue with this is the associated compound mediated cytotoxicity. As a consequence, it is necessary to determine to what extent any observation is related to the desired drug target and the undesired off-target effects that could underlie the observation.
With regard to cellular toxicity, cellular ATP is a commonly used marker for cell viability as this is the primary energy source in cells.3 Following the loss of membrane integrity during necrosis or the latter stages of apoptosis, cells are unable to synthesise ATP. Meanwhile, endogenous ATPases rapidly hydrolyse the remaining intracellular ATP. Therefore, the amount of cellular ATP positively correlates with the number of metabolically-active cells and a measure of cellular ATP can be used to quantify cellular toxicity. A particularly sensitive assay to measure compound-mediated cellular toxicity is the CellTiter-Glo®Luminescent Cell Viability Assay system. This utilises the available intracellular ATP by a reagent containing thermostable luciferase and luciferin, which undergoes oxidation to oxy-luciferin resulting in the production of light. The amount of light produced correlates with the number of metabolically-viable cells.
In a Proof-of-Concept study, we have screened the SCREEN-WELL® FDA v. 2.0 Approved Drug Library,4 which is a widely used set of approved and clinical compounds, in the CellTiter-Glo®Luminescent Cell Viability Assay and report a standardised assay protocol and the output of the study. In this study, we made use of the MCF-7 cell-line as it is a well-characterised breast cancer cell-line and a good model system for drug discovery research.5,6 The results are publicly available and will be particularly useful for those scientists engaged in drug repurposing.7 This approach to drug discovery can accelerate as it exploits existing drugs that have well-characterised bioactivity, safety and bioavailability information, and is optimal for compounds whose patents have expired.8
Materials
Reagents
- RPMI 1640, w/o L-Glutamine, w/o Phenol Red (Capricorn, #RPMI-XRXA)
- Dulbecco’s PBS (1x), w/o Ca & Mg, w/o Phenol Red (Capricorn, #PBS-1A)
- Trypsin-EDTA (Capricorn, #Try-1B)
- Penicillin/Streptomycin (100x) (Capricorn, #PS-B)
- L-Glutamine (Capricorn, #GLN-B)
- Fetal Bovine Serum, Collected in South America, VE = 500ml (Capricorn, #FBS-12A)
- CellTiter-Glo Luminescent Cell Viability Assay (Promega, #G7571)
- DMSO (Carl Roth, #HN47.1)
- SCREEN-WELL® FDA v. 2.0 Approved Drug Library (Enzo Life Sciences, #BML-284)
- MCF-7 cells (Sigma-Aldrich, #86012803-1VL)
Equipment
- 384 Well Polystyrene Cell Culture Microplates, white (Greiner Bio-One, #781073)
- Polystyrene Lids (Greiner Bio-One, #656161)
- CELLSTAR® Filter Cap Cell Culture Flasks (Greiner Bio-One, #660175)
- Echo Qualified 384-Well COC Source Microplate (Labcyte, #LP-0200)
- Echo 550 R Liquid Handler (Labcyte Inc., #E5XX-0801)
- EnVision plate reader (PerkinElmer Inc.,#Serial 1030266)
- Multidrop Combi (Thermo Scientific)
- HeraSafe or Incubator I 189 (Thermo Scientific)
- Scepter™ 2.0 Cell Counter (Merck Millipore, PHCC20060).
Procedure
On the first day compounds and controls are added into assay plates using Echo 550 R Liquid Handler.
a) Thereafter 100nL are transferred from the SCREEN-WELL® FDA v. 2.0 Approved Drug Library (2mM stocks) and 100nL from plates containing the controls (DMSO 100 percent v/v and medium) (Figure 1). All compounds and controls were stored at -20°C in Echo qualified microplates and, before addition to wells, equilibrated at room temperature for 30 min and spun down.
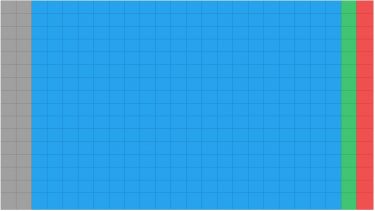

Figure 1. 384 well plate layout of the SCREEN-WELL® FDA v. 2.0 Approved Drug Library screen. grey = empty wells; blue = compound area; green = high control; red = low control.
b) MCF-7 cells were harvested from a 175cm2flask at 80 percent confluency by washing them once using 10mL room temperature PBS and afterwards incubating the cells with 5mL Trypsin 0.05 percent / EDTA 0.02 percent for 3 min. Cells are suspended using 10mL of pre-warmed cell culture media using RPMI-1640 supplemented with 10 percent FCS, 100U/mL Penicillin and 100µg/mL Streptomycin for MCF-7 cells respectively. Cell number is counted using a Scepter. Cells are diluted to a concentration of 50,000 cells per mL and 20µL of this suspension is added to each well of the cell culture microplate (1,000 cells/well) using the multidrop reagent dispenseror a multichannel pipette. Cells were incubated for an additional 24 h at 37°C, 5 percent CO2and humidified atmosphere.
To perform the CellTiter-Glo®Luminescent Cell Viability Assay the CellTiter-Glo® Buffer and lyophilised CellTiter-Glo® Substrate were thawed and left to equilibrate to room temperature prior to use.
a) 10mL of CellTiter-Glo®Buffer was transferred into the amber bottle containingCellTiter-Glo® Substrate to reconstitute the lyophilised enzyme/substrate mixture. The reagent was mixed by gently vortexing, swirling or inverting the contents to obtain a homogeneous solution and incubated at room temperature for 20 min. This formed the CellTiter-Glo® Reagent.
b) Assay plates containing the cells incubated with compounds were removed from the incubator and equilibrated for 30 min at room temperature.
c) 10µL of the CellTiter-Glo® Reagent was added to cells using a Multidrop reagent dispenser or multichannel pipette, assay plates were centrifuged for 1 min at 700 x g and incubated for 10 min in the dark at RT.
d) Luminescence was measured on the EnVision plate Reader (Table 1and Figure 2).
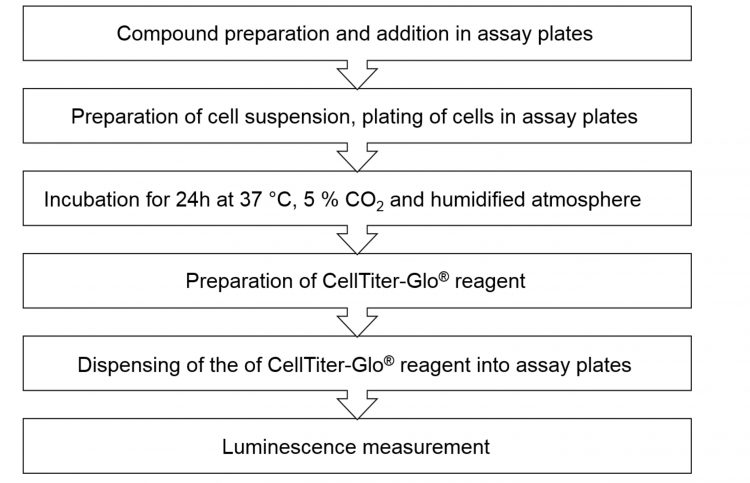

Figure 2: Workflow of the Proof-of-Concept screen.
The screening data was analysed using ActivityBase (IDBS), and outlier elimination in the controls was performed using the 3-sigma method. Compound data is normalised to the high (included 0.5 percent v/v DMSO – 100 percent viability) and low control (included medium only – 0 percent viability).
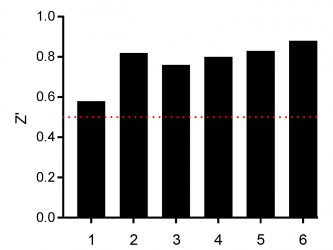

Figure 3: Determination of the Z’ values of screened assay plates with MCF-7 cells incubated for 24h for n=1 (plates 1-3) and n=2 (plates 4-6) with 10 µM compounds from the SCREEN-WELL® FDA v. 2.0 Approved Drug Library. The red dotted line is the minimum acceptable Z’ of 0.5.
Results & discussion
Before analysing the effect of the compound on cell viability, assay quality and robustness was analysed by calculating the Z’ value.8All assay plates yielded Z’ >0.5 revealing a robust and screening-compatible assay (Figure 3).
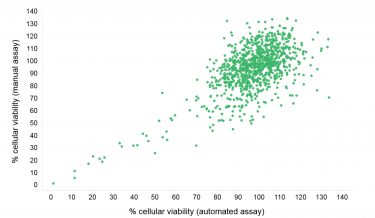

Figure 4: Compound effect on viability of MCF-7 cells seeded and screened automatically or manually. MCF-7 cells were incubated with compound for 24 h. Cell viability was subsequently detected using the CellTiter-Glo® Luminescent Cell Viability Assay that uses ATP as a marker for viability.
Cellular toxicity reproducibility was compared by performing the Proof-of-Concept screen in duplicate with the automated and manual dispensation of bulk reagents (Figure 4). A good correlation for the cellular toxicity was observed with most compounds appearing as non-toxic. A number of compounds were, however, seen to exhibit cellular toxicity to varying degrees. This exercise indicates that the CellTiter-Glo®Luminescent Cell Viability Assay is well suited for determining the cellular toxicity of compounds.
Biographies




References
- Zang R, Li D, Tang I-C, Wang J, Yang S-T. (2012). Cell-Based Assays in High-Throughput Screening for Drug Discovery. Int J Biotech Well Indus, 31-51.
- Michelini EC, Cevenini L, Mezzanotte L, Coppa A, Roda A. (2010). Cell-based assays: fuelling drug discovery. Anal Bioanal Chem, 227-238.
- Riss T. (2005). Selecting Cell based Assays for Drug Discovery Screening. Cell Notes Issue, 16-21.
- Kern FG, McLeskey SW, Zhang L, Kurebayashi J, Liu Y, Ding IY, Kharbanda S, Chen D, Miller D, Cullen K, Paik S, Dickson RB. (1994). Transfected MCF-7 cells as a model for breast-cancer progression. Breast Cancer Research and Treatment, 31, 153-165.
- Comşa Ş, Cîmpean AM, Raica M. (2015). The Story of MCF-7 Breast Cancer Cell Line: 40 years of Experience in Research. Anticancer Res, 35, 3147-3154.
- Enzo SCREEN-WELL ® COMPOUND LIBRARIES. Available online: http://www.enzolifesciences.com/fileadmin/files/catalog/Product-Guide_Compound_Libraries.pdf[Last accessed 07/11/2018].
- Pushpakom S, Iorio F, Eyers PA, Escott KJ, Hopper S, Wells A, Doig A, Guilliams T, Latimer J, McNamee C, Norris A, Sanseau P, Cavalla D, Pirmohamed M. (2018). Drug repurposing: progress, challenges and recommendations. Nat Rev Drug Discov. In press.
- Corsello SM, Bittker JA, Liu Z, Gould J, McCarren P, Hirschman JE, Johnston SE, Vrcic A, Wong B, Khan M, Asiedu J, Narayan R, Mader CC, Subramanian A, Golub TR. (2017). The Drug Repurposing Hub: a next-generation drug library and information resource. Nat Med, 405–408.
- Zhang JH, Chung TD, Oldenburg KR. (1999). A simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen2, 67-73.
Related topics
Assays, Cell-based assays, Drug Discovery, Screening, Toxicology
Related conditions
Breast cancer