Scientists unveil first molecular-level analysis of Omicron spike protein
Posted: 21 January 2022 | Victoria Rees (Drug Target Review) | No comments yet
Using cryo-electron microscopy, researchers have completed the world’s first molecular analysis of the SARS-CoV-2 Omicron spike protein.

Researchers at the University of British Columbia (UBC), Canada have conducted the world’s first molecular-level structural analysis of the SARS-CoV-2 Omicron variant spike (S) protein.
The analysis – done at near atomic resolution using cryo-electron microscopy – reveals how the heavily mutated Omicron variant attaches to and infects human cells.
“Understanding the molecular structure of the viral S protein is important as it will allow us to develop more effective treatments against Omicron and related variants in the future,” said lead author Dr Sriram Subramaniam. “By analysing the mechanisms by which the virus infects human cells, we can develop better treatments that disrupt that process and neutralise the virus.”
The S protein, which is located on the outside of a coronavirus, enables SARS-CoV-2 to enter human cells. The Omicron variant has an unprecedented 37 mutations on its S protein – three to five times more than previous variants.
The structural analysis, published in Science, revealed that several mutations (R493, S496 and R498) create new salt bridges and hydrogen bonds between the spike protein and the human cell receptor known as ACE2. The researchers concluded that these new bonds appear to increase binding affinity, while other mutations (K417N) decrease the strength of this bond.
“Overall, the findings show that Omicron has greater binding affinity than the original virus, with levels more comparable to what we see with the Delta variant,” said Subramaniam. “It is remarkable that the Omicron variant evolved to retain its ability to bind with human cells despite such extensive mutations.”
The researchers conducted further experiments showing that the Omicron S protein exhibits increased antibody evasion. In contrast to previous variants, Omicron showed measurable evasion from all six monoclonal antibodies tested, with complete escape from five. The variant also displayed increased evasion of antibodies collected from vaccinated individuals and unvaccinated COVID-19 patients.
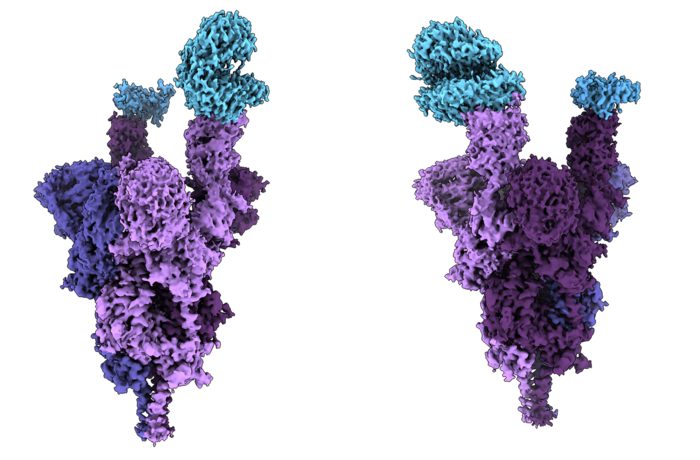

Atomic structure of the Omicron variant spike protein (purple) bound with the human ACE2 receptor (blue) [credit: UBC Faculty of Medicine].
“Notably, Omicron was less evasive of the immunity created by vaccines, compared to immunity from natural infection in unvaccinated patients. This suggests that vaccination remains our best defence,” said Subramaniam.
Based on the observed increase in binding affinity and antibody evasion, the researchers say that the S protein mutations are likely contributing factors to the increased transmissibility of the Omicron variant.
Next, Subramaniam says his research team will leverage this knowledge to support the development of more effective treatments.
“An important focus for our team is to better understand the binding of neutralising antibodies and treatments that will be effective across the entire range of variants and how those can be used to develop variant-resistant treatments,” Subramaniam concluded.
Related topics
Imaging, Microscopy, Protein, Proteomics, Structural Biology
Related conditions
Covid-19
Related organisations
University of British Columbia (UBC)
Related people
Dr Sriram Subramaniam