New study sheds light on drug-resistance mechanism
Posted: 25 February 2022 | Ria Kakkad (Drug Target Review) | No comments yet
MIT chemists have discovered a possible way to design drugs that could fight against drug resistance.


Researchers at the Massachusetts Institute of Technology (MIT), US have discovered the structure of a protein that can pump toxic molecules out of bacterial cells. Proteins like this one are believed to help bacteria become resistant to multiple antibiotics and the results of this study make it potentially possible to design drugs that could block these transport proteins and help drug-resistant bacteria react to existing antibiotics.
A strategy that bacteria uses to evade antibiotics is by pumping out drugs through their cell membranes. A team from the University of Wisconsin, US has been studying a membrane-bound protein called EmrE, which can transport many different toxic molecules, including herbicides and antimicrobial compounds.
A few years ago, Professor Mei Hong, senior author of the paper published in Nature Communications, developed a technique that allows researchers to use nuclear magnetic resonance (NMR) to measure the distances between fluorine probes and hydrogen atoms in proteins. This made it possible to determine the structure of a protein as it binds to a molecule that contains fluorine. After developing this technique, she teamed up with the University of Wisconsin to study EmrE.
The research group at the University of Wisconsin had already spent many years studying how EmrE transports a drug-like molecule, or ligand, across the phospholipid membrane. This ligand, known as F4-TPP+, is a tetrahedral molecule with four fluorine atoms attached to it, one at each corner.
Using this ligand with Hong’s new NMR technique, the researchers set out to determine an atomic-resolution structure of EmrE. It was already known that each EmrE molecule contains four transmembrane helices that are roughly parallel. Two EmrE molecules assemble into a dimer, so that eight transmembrane helices form inner walls that interact with the ligand as it moves through the channel. Previous studies have revealed the overall topology of the helices, but not of the protein side chains that extend into the channel interior, which are like arms that grab the ligand and help guide it through the channel.
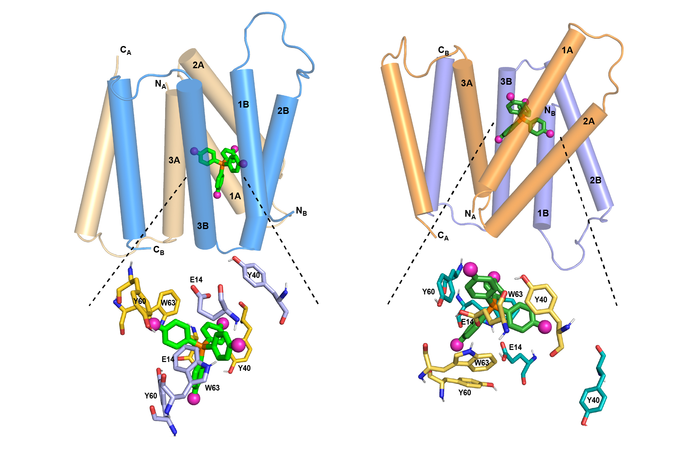

MIT chemists have discovered how the structure of the EmrE transporter changes as a compound moves through it. At left is the transporter structure at high pH. As the pH drops (right), the helices begin to tilt so that the channel is more open toward the outside of the cell, guiding the compound out.
[Credit: MIT]
EmrE transports toxic molecules from the inside of a bacterial cell, which is at neutral pH, to the outside, which is acidic. This change in pH across the membrane affects the structure of EmrE. In a 2021 paper, the researchers discovered the structure of the protein as it binds to F4-TPP+ in an acidic environment. In the new study, they analysed the structure at a neutral pH, allowing them to determine how the structure of the protein changes as the pH changes.
At neutral pH, the researchers found the four helices that make up the channel are relatively parallel to one another, creating an opening that the ligand can easily enter. As the pH drops, moving toward the outside of the membrane, the helices begin to tilt so that the channel is more open toward the outside of the cell. This helps to push the ligand out of the channel. At the same time, several rings found in the protein side chains shift their orientation in a way that also helps to guide the ligand out of the channel.
The EmrE channel is believed to transport many different toxic compounds. Therefore, the team now plans to study how other molecules travel through the channel.
Related topics
Drug Delivery, Drug Development, Drug Discovery, Drug Discovery Processes, Drug Targets
Related organisations
Massachusetts Institute of Technology (MIT), University of Wisconsin
Related people
Professor Mei Hong