AI for drug discovery in advanced pain management
Posted: 8 October 2024 | Drug Target Review | No comments yet
A deep-learning model found metabolites and drugs that could be repurposed for non-addictive and non-opioid options to treat chronic pain.
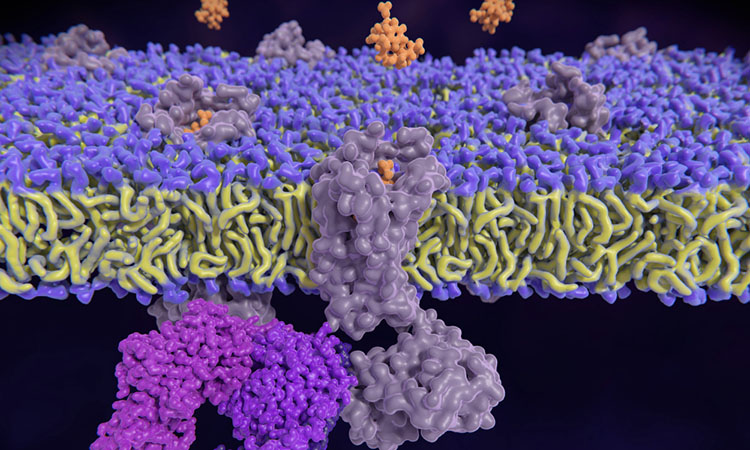

Researchers at Kent State University, Cleveland Clinic and IBM have created a deep-learning framework which identified multiple gut microbiome-derived metabolites and FDA-approved drugs that can be repurposed to select non-addictive, non-opioid options to treat chronic pain.
Chronic pain
An estimated one in five Americans live with chronic pain. However, treating chronic pain with opioids is difficult because of the risk of severe side effects and dependency. Recently, drugging a specific subset of pain receptors, G protein-coupled receptors (GPCRs), have shown evidence to be a non-addictive and non-opioid pain relief.
The team sought to apply research methods they had already developed for finding pre-existing FDA-approved drugs for potential pain indication, which involved mapping out gut metabolites to identify drug targets. A 3D understanding of molecules based on extensive 2D data about their physical, structural and chemical properties was required to determine whether a molecule will work as a drug.
Dr Feixiong Cheng, Director of Cleveland Clinic’s Genome Center, and IBM commented: “Even with the help of current computational methods, combining the amount of data we need for our predictive analyses is extremely complex and time-consuming…AI can rapidly make full use of both compound and protein data gained from imaging, evolutionary and chemical experiments to predict which compound has the best chance of influencing our pain receptors in the right way.”
LISA-CPI
The tool, named LISA-CPI (Ligand Image- and receptor’s three-dimensional (3D) Structures-Aware framework to predict Compound-Protein Interactions) uses deep learning to predict if a molecule can bind to a specific pain receptor, where on the receptor a molecule will physically attach, how strongly the molecule will attach to that receptor, and whether binding a molecule to a receptor will switch signalling effects turn on or off.
LISA-CPI was used to predict how 369 gut microbial metabolites and 2,308 FDA- approved drugs would interact with 13 pain-associated receptors and found numerous compounds that could be repurposed to treat pain. Currently, studies are validating these compounds in the lab.
Dr Yuxin Yang, a former Kent State University graduate student and current data scientist in Dr Cheng’s lab, explained: “This algorithm’s predictions can lessen the experimental burden researchers must overcome to even come up with a list of candidate drugs for further testing…We can use this tool to test even more drugs, metabolites, GPCRs and other receptors to find therapeutics that treat diseases beyond pain, like Alzheimer’s disease.”
This is one example of how the team is collaborating with IBM to develop small molecule foundation models for drug development.
This study was published in Cell Press.
Related topics
Artificial Intelligence, Drug Repurposing, GPCRs
Related conditions
Chronic pain
Related organisations
Cleveland Clinic, IBM, Kent State University