New study reveals how bacteria ‘vaccinate’ with viral DNA
Posted: 1 April 2025 | Drug Target Review | No comments yet
The discovery from researchers at Johns Hopkins Medicine reveals how bacteria use the CRISPR-Cas system to store viral DNA, enhancing their immunity against future infections, and potentially paving the way for new phage-based therapies
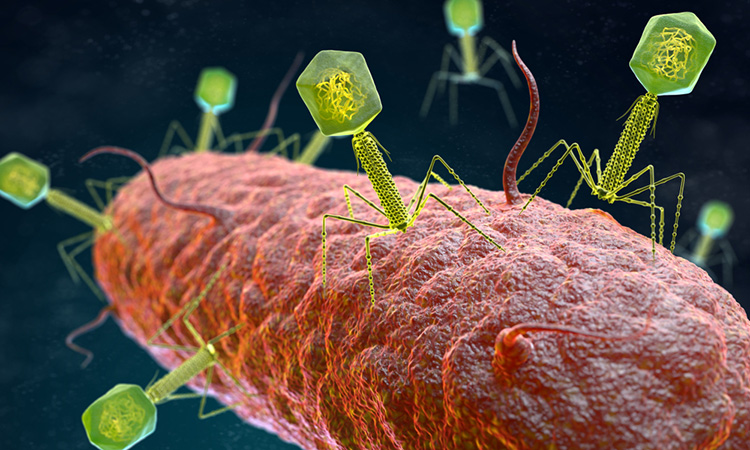

Like humans, bacteria are constantly under attack by viruses. These bacterial invaders, known as bacteriophages or “phages,” can either destroy bacterial cells or remain dormant within them. Scientists have long studied how bacteria survive phage infections to better understand immunity and develop new disease-fighting strategies.
Now, researchers at Johns Hopkins Medicine have uncovered a bacterial defence mechanism: bacteria can use genetic material from weakened, dormant phages to “vaccinate” themselves, equipping future generations with the ability to recognise and combat similar threats. Their findings, published in Cell Host & Microbe, provide new insights into bacterial immunity and could inform the development of novel phage-based therapies for antibiotic-resistant infections.
How bacteria ‘remember’ their viral enemies
The research team, led by Dr Joshua Modell, focused on Streptococcus pyogenes, the bacteria responsible for strep throat. They studied a class of phages called temperate phages, which can either kill bacterial cells or enter a dormant state. During this dormancy, bacteria steal genetic fragments from the phages and incorporate them into their own genome. This genetic memory is passed down to offspring, allowing them to recognise and fight off future infections from the same phage.
“This is conceptually similar to a vaccine with an attenuated virus,” explains Nicholas Keith, a graduate student and first author of the study.
CRISPR: bacteria’s built-in defence system
Bacteria rely on the CRISPR-Cas system to protect themselves from viral attacks. This system acts as a molecular recording device, capturing snippets of viral DNA from previous infections. When a bacterium encounters the same virus again, CRISPR-Cas recognises the invader and destroys its genetic material.
When a bacterium encounters the same virus again, CRISPR-Cas recognises the invader and destroys its genetic material.
To test how bacteria acquire these genetic memories, researchers infected bacterial populations with both naturally occurring temperate phages and genetically engineered non-dormant phages. Their results showed that bacteria were much more effective at capturing viral DNA from dormant phages than from active ones. Using genome sequencing, they identified hundreds of thousands of new DNA memories stored by the CRISPR-Cas system.
“Our results indicate that the bacteria’s CRISPR system was more effective at using the naturally dormant phage to pull parts of the viral genetic code into their genome,” says Modell. “When we tested phages that could not go dormant, the CRISPR system did not work nearly as well.”
Implications for human health
The study helps scientists understand how bacteria acquire toxic genes from phages, these are genes that can make infections such as Staphylococcus aureus (Staph), Escherichia coli (E. coli), and cholera more dangerous to humans. Understanding bacterial immunity could also aid in the design of phage therapies, which use viruses to target antibiotic-resistant bacteria.
Modell explains: “CRISPR systems are one of the first lines of defence against hazardous genes from phages that turn bacterial cells toxic; furthermore, our studies will inform the design of phage therapies that could be used in clinical cases where a bacterial infection is resistant to all available antibiotics.”
Future research will focus on how CRISPR systems protect bacteria from viruses that do not enter a dormant state, potentially leading to new medical and biotechnological applications.
The study was conducted by researchers at Johns Hopkins University School of Medicine, Hunter College, and the National Institutes of Health, with funding from the NIH, the Rita Allen Foundation, and the National Science Foundation.
Related topics
Bacteriophages, Genetic Analysis, Genome Editing, Genomics, Microbiology, Virology
Related conditions
bacterial infections, Cholera, Escherichia coli, Staphylococcus aureus
Related organisations
Hunter College, John Hopkins Medicine, National Institutes of Health (NIH), National Science Foundation, The Rita Allen Foundation
Related people
Dr Joshua Modell