Scientists develop novel method for membrane protein crystallisation
Posted: 8 March 2018 | Drug Target Review | No comments yet
Scientists in Switzerland have developed a new means of crystallising membrane proteins – a hitherto elusive feat – in order to study their structure and aid understanding of their processes.

Membrane-embedded proteins are an essential part of cells and any form of life. They not only exist in many different forms, but also perform a wide range of functions – ranging from intracellular communication and the transport of substances into or out of the cell, to mediating the immune response. Membrane proteins are considered important therapeutic and diagnostic target structures. If their structure and functions are known, pharmaceutical researchers can develop active substances that influence these functions in a targeted manner.
Until now, however, elucidating the structure of membrane proteins has been very difficult since it first requires researchers to isolate large numbers of these molecules and form crystals from them. Therein lies the difficulty: membrane proteins are insoluble in water and often too large and heterogeneous to be crystallised using standard methods.
Now, the group led by Raffaele Mezzenga, Professor of Soft Materials at ETH Zurich, is working to eliminate this restriction by developing a general method that can be used to crystallise membrane proteins of any type or size.
Lipidic mesophases enable crystallisation
Their method is built upon a technique used in the 1990s called ‘in meso crystallisation’, where proteins are isolated and concentrated using stable water-lipid mixtures known as lipidic mesophases.
In mesophases of this kind, a self-assembly process creates a three-dimensional network of bent water channels whose walls are made up of lipids, as in a biomembrane. These water channels typically have a diameter of 3-4 nanometres, and the network’s basic cubic motif is repeated at regular intervals.
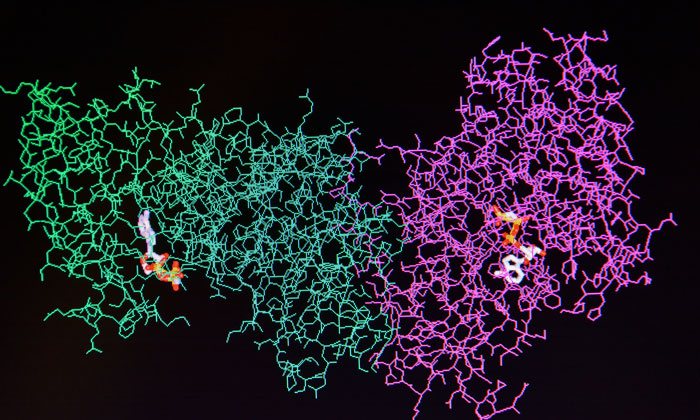
In channels of this kind, the membrane proteins embed themselves into the walls using the hydrophobic part that otherwise sits in the cell membrane. The rest of the protein ends up in the water channel’s interior, and the proteins, once correctly reconstituted, can then start to crystallise. The fact that the channels offer so little space, has meant that previously only small membrane proteins could be crystallised – large proteins were crushed out and did not form crystals.
Making room for large membrane proteins
In order to accommodate larger membrane proteins, ETH researchers use a trick to expand the channels. By mixing a small proportion of electrically charged lipids in with the lipids, they repel one another and thus inflate the channels, increasing their diameter to 20 nanometres. This is the first demonstrated evolution of attempts to electrostatically swell these water channels into a methodology of general significance.
Owing to these swollen lipidic mesophases, Mezzenga and his colleagues managed to crystallise large membrane proteins and consequently elucidate their structure.
Structural biologists and pharma to gain from method
The new ‘generalised in meso’ method is likely to be of great interest to structural biologists in particular, who until now have struggled to elucidate the structure of large membrane proteins.
“This tool will give new impetus to structural elucidation, as it opens up proteins that were previously out of reach,” says Mezzenga.
At present, scientists know the precise structure of only 360 small membrane proteins, or about a seventh of all membrane proteins. The structure of the many remaining membrane proteins is unknown.
According to Mezzenga, the research may also be of benefit to the pharmaceutical industry. “The ability to determine structure is of paramount importance for the development of new drugs,” he says. “This method will make it considerably easier and provide new impetus in the field.”The development is not covered by a patent. “We purposely published the paper in an open access journal, so that all interested researchers have unlimited access to it,” says Mezzenga, who has invested more than three years of work in the project.
Details of this new method were published in the journal Nature Communications.
Related topics
Crystallography, Imaging, Molecular Biology
Related organisations
ETH Zurich
Related people
Prof Raffaele Mezzenga