New ligand binding site identified through computer simulations
Posted: 30 July 2018 | Drug Target Review | No comments yet
Many therapeutic compounds target GPCRs and so new ligand binding sites can help to design drugs more efficient at targeting GPCR…
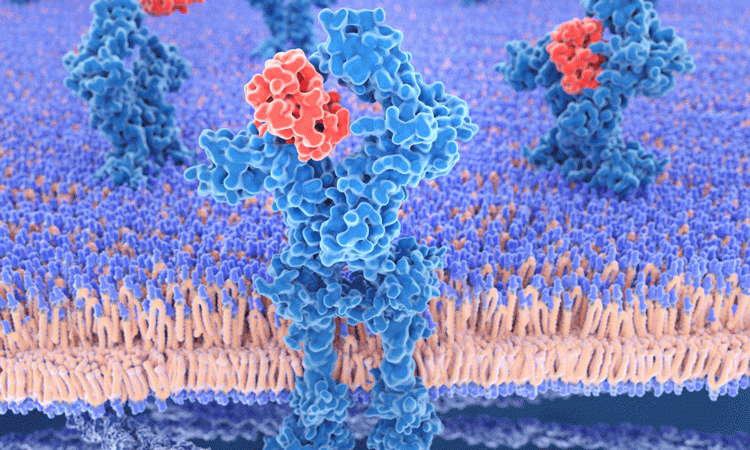

A novel binding site for natural ligands and drugs has been identified by researchers using a computational simulation.
EPFL scientists used a simulation of an important receptor to identify the site, which may be present on other receptors and can be used in new treatments for various diseases.
A receptor is a specialised protein which is activated when a ligand binds to it. Ligands are molecules which bind to the target site, and can be various substances, hormones, nucleic acids or neurotransmitters. By binding proteins, ligands run complex processes such as immune responses, cell development and genetics. Most biological processes occur through a receptor, and as such is vital.
A team of scientists led by Professor Horst Vogel at EPFL developed a computer simulation of the widespread muscarinic acetylcholine receptor, examining the M3 and M4 subtypes of the receptor, which are involved in the function of the lungs and the central nervous system.
These receptors belong to a family of G protein-coupled receptors (GPCRs) which detect signals coming from outside the cells, such as hormones, light or neurotransmitters. Once they are activated, GPCRs change their structure so they can bind and activate other proteins inside the cell and stimulate the appropriate process.
Many therapeutic compounds target GPCRs and so new ligand binding sites can help to design drugs more efficient at targeting GPCR.
Molecular dynamics simulations were used to study the physical movement of atoms and molecules within the protein on a computer. This method reveals minute details, offering a high resolution method of identifying methods and locations of ligand binding.
Researchers found a new binding site on acetylcholine receptors, which can be pharmacologically exploited in order to understand the process of ligand binding and activation. The identified how the entire site expands when a ligand binds to a new site, and how the site seemed capable of binding small ligands and causing different effects to the receptor than the ‘main’ ligand would.
The team looked at more than 200 ligand-bound GPCR structures in this way and discovered that most ligands bind to the orthosteric sites on GPCR. However, one receptor that binds to leukotriene (LTB4) and directs immune cells to site of infection seemed to bind a ‘double’ ligand – one which binds to the new site in the two acetylcholine receptors. The new site was detected in many of the other 200 receptors studied.
This new binding site may exist in other GPCRs, which opens an opportunity for GPCR drug discovery.
Prof Vogel said, “The study shows the power of computational methods to resolve in atomic detail central receptor-mediated signalling reactions.
The challenging next step is to use computational methods to design novel compounds that would fit into the newly found binding sites to activate or deactivate the receptor in a defined mode and thus design novel medicines.”
Related topics
Bioinformatics, Drug Discovery, Research & Development, Target Molecule
Related organisations
Ecole Polytechnique Fédérale De Lausanne (EPFL)
Related people
Professor Horst Vogel