Targets for chronic kidney disease treatment
Posted: 2 October 2018 | Iqra Farooq (Drug Target Review) | No comments yet
The DAB2 pathway could be a crucial target in treating CKD, as mouse models show a decrease in DAB2 leads to prevention of the condition…

DAB2 could be a target for chronic kidney disease treatments after researchers investigated how genetic variations drive the expression of genes within the cells of the kidney.
The kidney works harder than any other organ in the body, tasked with extracting waste, balancing body fluids, forming urine, regulating blood pressure and secreting hormones. When the kidney goes wrong, the body’s systems can be put in danger from toxin accumulation, fatigue and high blood pressure.
Researchers from the Perelman School of Medicine at the University of Pennsylvania investigated how genetic differences drive the varying expression of genes within the filtering cells of the kidney. They identified new pathways that explained chronic kidney disease (CKD) development.
CKD affects over 700 million people around the world, with approximately 14 percent of the American population affected by the condition.
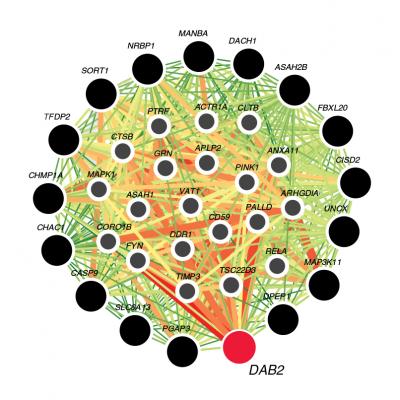

Visualization of tubule-specific candidate genes. The color of each link shows the relationship confidence, from green (0.01) to red (1). Credit: Katalin Susztak, MD, PhD, University of Pennsylvania.
The study was led by Dr Katalin Susztak, Professor of Renal-Electrolyte and Hypertension and Genetics at the Perelman School of Medicine.
Previously, Prof Susztak’s lab had generated an atlas for the kidney, which included a novel molecular definition of all cell types in the kidney. They identified that each distinct type has a unique function, and specific dysfunction is associated with specific symptoms in those with CKD.
“This study is the first to look at specific cell types and how their genetic variations can lead to disease development,” Prof Susztak said.
The team developed a database to show how genetic variation influences mRNA expression in kidney cells. They integrated this information, with that from genome-wide association analysis (GWAS) and more specific approaches to identify genes associated with CKD.
“In the past, many GWAS efforts have identified sequence variants for CKD, but the biological basis of these variants was poorly understood,” Prof Susztak said. “We need to do more with all of the information we have sitting in GWAS databases to identify the genes, cells, and molecular pathways responsible for CKD.”
The researchers identified that genes though to be associated with CKD were more abundant in the proximal tubule of the kidney. From these 27 genes, they focused on the DAB2 gene from the TGF-β pathway, and found that it connected to many other genes central to proper kidney function.
Experiments using mouse models confirmed that the reduction of DAB2 meant the mice were protected from CKD. Lowering the levels of DAB2 led to the cytokine TGF-β pathway not inducing fibrosis as a healing reaction.
“We are just starting to find which molecules have gone astray to cause disease in order to develop drugs to counteract overactive molecules that cause damage to healthy tissue,” concluded Prof Susztak.
The study was reported in Nature Medicine.
Related topics
Disease Research, Drug Discovery, Drug Targets
Related conditions
Chronic kidney disease
Related organisations
Perelman School of Medicine at the University of Pennsylvania
Related people
Professor Katalin Susztak








