Digital 3D brain cell atlas released
Posted: 30 November 2018 | Fraser Owen (Drug Target Review) | No comments yet
The Blue Brain 3D atlas could be the future of analysing all cells in the brain, with high quality, high resolution images that can navigate the brain…
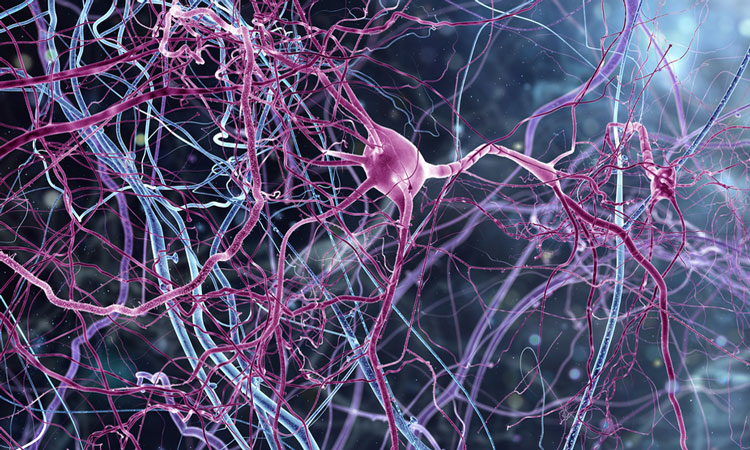

The first digital 3D atlas of every cell in the mouse brain provides neuroscientists with previously unavailable information on major cell types, numbers, and positions of all 737 brain regions – which will potentially accelerate progress in brain science massively. Released by EPFL’s Blue Brain Project, the Blue Brain Cell Atlas integrates data from thousands of whole brain tissue stains into a comprehensive, interactive and dynamic online resource that can continuously be updated with new findings. This groundbreaking digital atlas can be used for analysing and further modeling specific brain areas, and is a major step toward a full simulation of the rodent brain.
“Despite vast numbers of studies over the past century, cell numbers were still only available for 4 percent of mouse brain regions – and these estimates often varied by as much as three-fold,” said Blue Brain Project Founder and Director, Professor Henry Markram. “The Blue Brain Cell Atlas solves this problem by presenting the best estimates for even the smallest-known region of the mouse brain.”
“Knowing the circuit components and how they are arranged is also an essential starting point for modelling the brain – just as demographic data are essential for modelling a country, for example,” explained lead author and creator of the Cell Atlas, Dr Csaba Erö.
Biomarkers aren’t just supporting drug discovery – they’re driving it
FREE market report
From smarter trials to faster insights, this report unpacks the science, strategy and real-world impact behind the next generation of precision therapies.
What you’ll unlock:
- How biomarkers are guiding dose selection and early efficacy decisions in complex trials
- Why multi-omics, liquid biopsy and digital tools are redefining the discovery process
- What makes lab data regulatory-ready and why alignment matters from day one
Explore how biomarkers are shaping early drug development
Access the full report – it’s free!
Previous brain atlases consist of stacks of images of stained brain slices. Some show precise cell positions for the entire brain, while others show particular cell types – but none turns this valuable data into numbers and positions of all the cells in the brain in the form of a digitally navigable atlas.
This revolutionary step took five years of carefully collecting and integrating thousands of stainings of brain tissue. Dr Erö and colleagues relied mostly on all the imaging data made available from the Allen Institute for Brain Science and combined this with a large number of other anatomical studies to calculate and validate the major types, numbers, and positions of cells in each area of the mouse brain – including all the regions where cell data was never obtained before.
“Our Cell Atlas is like going from hand-drawn maps to digitised versions of satellite images of cities and geographical features – allowing us to navigate the brain the way Google Earth allows us to navigate the Earth,” said Blue Brain Section Manager, Dr Marc-Oliver Gewaltig. “It’s 3D, it’s high resolution, it’s searchable, it’s navigable, it’s annotated, it’s user-friendly – and it fills a huge gap in our knowledge of 96 percent of the mouse’s brain regions.”
Freely available online, the Blue Brain Cell Atlas allows users to visualise all 737 brain regions and the cells they contain, and to download the region with their numbers and locations. It distinguishes excitatory, inhibitory and some other types of neurons – as well as major types of non-neuronal cells, called glia, which insulates and protect neurons. These data are important for researchers trying to understand the structure and function of different brain regions or for building functional models of specific brain regions.
“It is also a great teaching aid: you can choose to display just the regions of interest and navigate through these down to the scale of individual cells, which are color-coded by the type of their morphology,” added Dr Gewaltig.
The Blue Brain Cell Atlas is also the first brain atlas that is dynamic — allowing researchers to contribute to and improve the atlas with any new data. “We can now move collaboratively towards the ground truth of what is inside the mouse brain,” explained Prof Markram.
“Our message to brain researchers everywhere is: try it, use it, add data to it,” concluded the authors.
The study was published in Frontiers in Neuroinformatics.
Related topics
Analysis, Analytical Techniques, Big Data, Bioinformatics, Disease Research, Genomics, Research & Development, Therapeutics
Related organisations
Allen Institute for Brain Science, EPFL's Blue Brain Project
Related people
Dr Csaba Erö, Dr Marc-Oliver Gewaltig, Professor Henry Markram