CRISPR repurposed to develop better antibiotics
Posted: 14 January 2019 | Iqra Farooq (Drug Target Review) | No comments yet
Mobile-CRISPRi could be used to guide gene expression, in turn control levels of proteins produced, enabling scientists to identify key antibiotic targets…
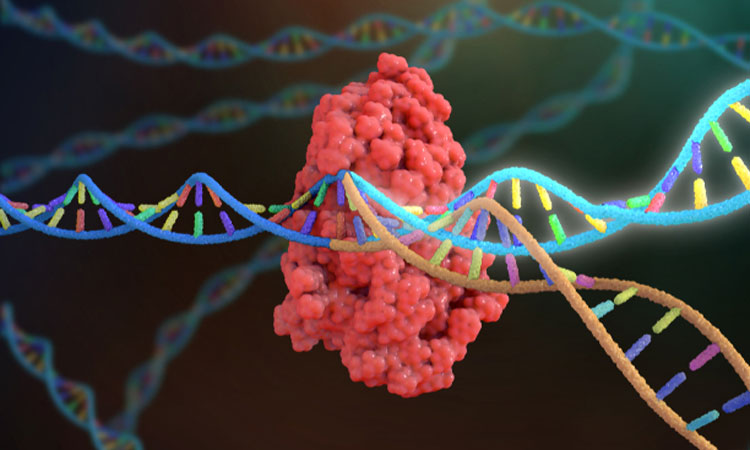

A team of researchers have successfully repurposed the gene editing CRISPR tool to develop better antibiotics, calling the technique Mobile-CRISPRi.
The team of researchers, at the University of Wisconsin-Madison collaborated with researchers at the University of California, San Francisco, to use the technique to study which genes are targeted by particular antibiotics, and using the information to improve current antibiotic drugs, or to develop new ones.
“What we need to do is to figure out new weaknesses in these bacteria,” said Professor Jason Peters, a UW-Madison Professor of Pharmaceutical Sciences, who developed the new system.
Biomarkers are redefining how precision therapies are discovered, validated and delivered.
This exclusive expert-led report reveals how leading teams are using biomarker science to drive faster insights, cleaner data and more targeted treatments – from discovery to diagnostics.
Inside the report:
- How leading organisations are reshaping strategy with biomarker-led approaches
- Better tools for real-time decision-making – turning complex data into faster insights
- Global standardisation and assay sensitivity – what it takes to scale across networks
Discover how biomarker science is addressing the biggest hurdles in drug discovery, translational research and precision medicine – access your free copy today
Resistance to current antibiotics is now a growing problem, with estimates suggesting that millions of lives could be endangered, costing the US $2 billion each year.
The technique allows the scientists to screen for antibiotic function in a wide range of pathogenic bacteria.
The team transferred Mobile-CRISPRi from common laboratory strains into diverse bacteria, even including a little-studied microbe making its home on cheese rinds. The researchers mentioned how the ease of the transfer made the technique a favourable option when studying bacteria that cause disease or promote health.
The system reduces the production of protein from targeted genes, and allowed the researchers to identify how antibiotics inhibit the growth of pathogens.
“Most people, when they think about CRISPR, think about gene editing,” said Prof Peters, who earned his doctorate at UW-Madison and joined the School of Pharmacy as an Assistant Professor. “But that’s not what I do.”
Ordinarily, CRISPR would cut the DNA into two, the gene could then be edited, while the cell repairs the damage.
The researchers instead used CRISPRi, a defanged form of the technique that is unable to cut the DNA, but sits and deflects other proteins, preventing them from gaining access to particular genes to turn then on.
This results in a lower expression of the gene, and thus a reduced amount of the protein it codes for. The mentioned how if they decreased the amount of protein targeted by an antibiotic, bacteria became much more sensitive to lower levels of the drug, adding to evidence of an association between gene and drug.
In this way, thousands of genes can be screened as potential antibiotic targets, help the understanding of how antibiotics work and how to improve them.
The make the technique mobile, the team developed methods to transfer the system from common lab models like E. coli to disease-causing species, which are often harder to study. They implemented a method called conjugation.
“You basically mix the bacteria together and it happens,” Prof Peters said of conjugation. “It doesn’t get much easier than that.”
The team transferred Mobile-CRISPRi using conjugation to the pathogens Pseudomonas, Salmonella, Staphylococcus and Listeria, and some others.
“What that means is that you can now do studies on how antibiotics work directly in these pathogens,” said Prof Peters. “That could give us a better clue about how these drugs work in the different organisms and potentially what we can do to make them better.”
“So now it’s going to be completely available to the community,” said Prof Peters. “Now this gives people a path forward.”
The findings of the study were published in the journal Nature Microbiology.
Related topics
Antibiotics, CRISPR, Disease Research, Drug Discovery, Drug Targets, Genomics, Research & Development, Screening, Therapeutics
Related organisations
California University, University of Wisconsin-Madison
Related people
Professor Jason Peters