Key mechanism in tuberculosis survival discovered
Posted: 19 August 2019 | Victoria Rees (Drug Target Review) | No comments yet
A new study has revealed that M. tuberculosis uses a unique type of antacid which gives immune cells indigestion, enabling the bacteria to survive.

A study has discovered a key mechanism the Mycobacterium tuberculosis (TB) bacteria uses that prevents immune cells from killing them. According to the researchers, their findings could be used to develop novel treatments to combat the condition.
The investigation was conducted by Harvard Medical School, US, with a collaborative team including researchers from the University of Groningen, The Netherlands. Previous studies have shown that M. tuberculosis can survive for years despite encapsulation inside phagosomes within macrophages. This means that antibiotics are not always able to be effective.
An investigation led by the team in 2014 identified lipids present in M. tuberculosis but not in M. bovis, a less harmful bacterium. They concluded that the lipids in M. tuberculosis could therefore be important in virulence and found a candidate, called 1-tuberculosinyladenosine (1-TbAd), an adenosine modified by the attachment of a lipid in the 1-position.
“Such a modification is extremely rare in nature,” said Dr Jeffrey Buter, first author of the paper, “yet M. tuberculosis produces and releases a relatively large amount of this compound.”
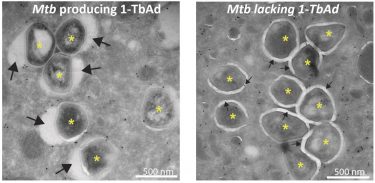

Electron microscopy pictures showing the bacteria producing 1-TbAd are surrounded by a swollen phagosome (left picture) and bacteria not producing 1-TbAd are surrounded by tight phagosomes (right picture) [credit: Buter et al/Nature Chemical Biology].
They uncovered two enzymes critical to the production of 1-TbAd, but not the mechanism by which the molecule helped the bacterium survive. Discovering research conducted by another group, the team learned that the fusion of the phagosome and lysosome was blocked due to these enzymes. As the phagosome needs to be acidic for fusion, the researchers hypothesised that 1-TbAd played a role in preventing acidification of the phagosome.
“1-TbAd is a weak acid and in equilibrium with its basic counterpart,” said Buter. “In the acidic environment of the phagosome, this base will reduce acidity.” This suggests that the molecule works as an antacid and prevents the phagosome from reaching the level of acidity required to fuse with the lysosome.
The researchers investigated the adenosine compound, to understand whether it acts through a receptor. They confirmed that 1-TbAd works by reducing the acidity directly, so synthesised different variants of the molecule to determine which parts were vital to its function.
“The lipid part is needed to cross the membranes and get into the phagosomes and lysosomes,” explained Buter.
Further microscopy studies performed in the lab of Nicole van der Wel at the Amsterdam UMC reveal that exposing macrophages to 1-TbAd makes their lysosomes swell up to five times their normal size. Tests with macrophages infected with M. tuberculosis show that the phagosomes only increase significantly in the presence of the enzyme Rv3378c needed to produce 1-TbAd.
Additionally, the mechanism by which 1-TbAd works is the same as how chloroquine kills the malaria parasite, which blocks the functioning of parasite’s lysosomes. This suggests that 1-TbAd could be used as an anti-malarial drug but also targeting the production of 1-TbAd could kill M. tuberculosis inside the macrophages. The researchers say that the enzyme Rv3378c would be an interesting target for drug discovery, as it is unique to the tuberculosis bacterium.
The findings were published in Nature Chemical Biology.
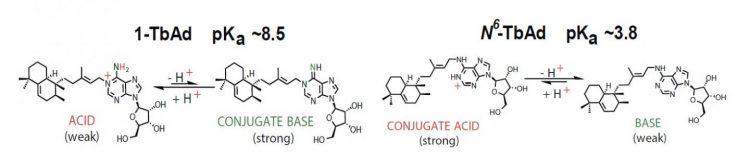

1-TbAd acts as an acid or conjugated base depending on the acidity of the environment [credit: Buter et al/Nature Chemical Biology].
Related topics
Drug Discovery, Drug Targets, Enzymes, Research & Development
Related conditions
Tuberculosis (TB)
Related organisations
Harvard Medical School, Nature Chemical Biology, University of Groningen
Related people
Dr Jeffrey Buter