Faster drug discovery could hit ‘undruggable’ targets
Posted: 23 January 2020 | Rachael Harper (Drug Target Review) | No comments yet
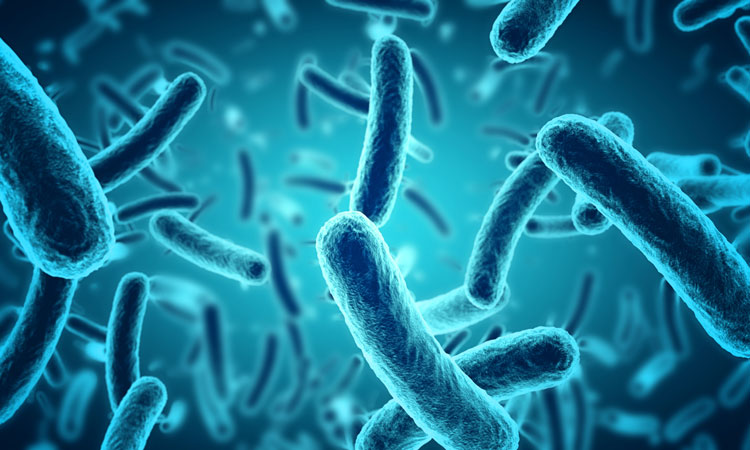
A new technique has been developed which has the potential to harness bacteria to produce billions of different drug candidates to treat diseases like cancer.

Medicines made from coiled protein fragments could provide a new handle on hard-to-treat diseases like cancer; now, a new technique has been developed which has the potential to harness bacteria to produce billions of different drug candidates that will not fall apart quickly inside the body. The technique was developed by the University of Michigan (UM), US.
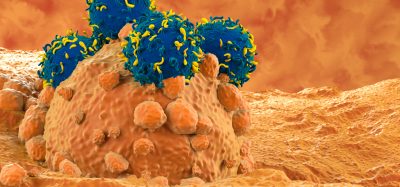
Some advanced cancer medications target cancer cells with immune markers that latch onto the outsides of the cancer cells. These immune markers are large, so they operate between cells rather than inside them. To disrupt cancer cells from within, the medicine must be small enough to get into the cell.
However, ordinary small-molecule medications cannot hit messenger proteins with smooth surfaces; a new type of medication is needed.
“It’s a Goldilocks principle,” said Greg Thurber, associate professor of chemical engineering at UM, who led the work. “The drug has to be small enough to get into cells but large enough to grab onto proteins inside cells that don’t interact with ordinary small-molecule drugs.”
Protein fragments, known as peptides, could fill this gap. One example of an ‘undruggable’ target is MDM2, a protein that sidelines another protein known as p53, which stops cell division when genetic mutations arise, buying time for DNA repair and is the front-line defence against cancer.
However, cancer cells fight back by overproducing p53’s inhibitor, MDM2, which binds to p53 and prevents it from doing its job. If something intercepts the MDM2 before it ties up the p53, though, then the cancer cells might self-destruct.
This is just one of the therapies that might be possible with peptides and Thurber’s team has now stabilised them. “The peptide is like a Slinky, but you tie together links so it doesn’t stretch out. That’s what we do chemically,” said Thurber.
The tie enables different drug candidate peptides to be built with bacteria which are so plentiful that researchers can see how well they work right on the bacterium.
Thurber’s team used E. coli bacteria to build hundreds of millions of peptides with random variations, looking for the one that was best at binding MDM2. The instructions for the peptides were written into rings of genetic code that the researchers slipped into the bacteria, causing the bacteria to produce the peptides on their cell membranes.
The ties were made from synthetic molecules that the bacteria swapped in for a peptide building block that they could not produce. Unlike earlier ties incorporated into phage-built peptides, the synthetic molecules will not bind to other molecules in and around the bacteria.
The tie enables different drug candidate peptides to be built with bacteria”
“The problem was that the reaction to tie the coils together killed the bacteria,” said Tejas Navaratna, a PhD student in chemical engineering at UM and first author of the study. “We spent months trying to optimise the reaction so that the bacteria survived, but we eventually realised we’d have to extract the DNA that coded our best peptides and add it to fresh, live bacteria.”
Each bacterium produced a different peptide, enabling the team to test hundreds of millions of different designs. To find out whether the peptides worked, the researchers mixed MDM2 – attached to fluorescent molecules – in with the bacteria. The researchers concluded that the cells which lit up were producing a potentially useful drug.
The study was published in the Journal of the American Chemical Society.
Related topics
Drug Development, Drug Discovery Processes, Drug Targets, Peptide Therapeutics
Related conditions
Cancer
Related organisations
Michigan University
Related people
Greg Thurber, Tejas Navaratna