Imaging technique reveals how HIV drugs bind to viral structure
Posted: 31 January 2020 | Victoria Rees (Drug Target Review) | No comments yet
A new study has shown in three dimensions how drugs bind to HIV, which could be used to improve the design of drugs or develop novel therapies.

Researchers have discovered how a powerful class of HIV drugs binds to a key piece of the condition’s machinery. By solving, for the first time, three-dimensional (3D) structures of this complex while different drugs were attached, the scientists showed what makes the therapy so potent.
According to the researchers, from the Salk Institute, US, their findings provide insights that could help design or improve new treatments for HIV.
“The drugs we studied are the latest compounds available in the clinic today, as well as several important pre-clinical molecules. Until now, no one knew exactly how they bound to this HIV complex,” says the study’s senior author Dmitry Lyumkis, an assistant professor in Salk’s Laboratory of Genetics.
Drugs called integrase strand transfer inhibitors (INSTIs) block the intasome, a crucial structure of the virus that enables infection; HIV cannot infect human cells when the complex is not able to integrate viral DNA into the human genome.
Despite the success of these molecules, researchers have struggled to study how they inhibit the HIV intasome, largely due to difficulty in isolating intasomes for structural studies. In the past, most research on the intasome and INSTIs was carried out on another retrovirus called prototype foamy virus (PFV). In 2017, Lyumkis and his colleagues were the first to determine the structure of purified HIV intasomes.
In the new work, the team obtained the structure of HIV intasomes while being actively blocked by one of four INSTIs, the commercially available drug bictegravir or three experimental compounds known as 4f, 4d and 4c. The group used tilted single-particle cryo-electron microscopy (cryo-EM) to reveal the structure of each intasome-drug complex.
Binding to HIV
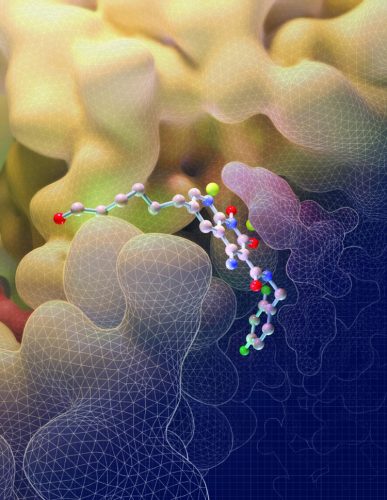

This illustration depicts the molecular structure of an HIV drug, known as an INSTI, binding to key sites on the intasome (yellow), the viral machine that allows HIV to invade cells (credit: Salk Institute).
The first observation made was how differently the drugs attached to the HIV intasome than to the PFV intasome. The compound known as 4f, for instance, loops backwards onto itself as it binds to the PFV intasome but remains relatively flat as it attaches to the HIV version of the complex, details which can help researchers improve the binding properties of potential future molecules.
“To this day, everyone is still using the PFV intasome structure to rationalise and understand the mechanism of action of these drugs,” says Dario Passos, the study’s co-first author. “But we’ve shown that the field really needs to move and study the HIV structure if we want to make further progress.”
The structures also revealed why the drugs are so potent and what makes them so good at avoiding drug resistance. The INSTIs fill the entire space that is normally occupied by DNA. That means if the HIV intasome develops a mutation that blocks the INSTI drugs from binding, it also blocks the DNA from attaching, rendering the complex useless for invading human cells.
Finally, the extremely high resolution of the structure obtained by the Salk researchers allows them to see details on how the drugs chemically interacted with this binding pocket and how INSTIs displaced water molecules to do so, which gave the team even more information on what makes INSTIs so successful in the clinic.
“In previous structures, we learned about intasome biology,” says Lyumkis. “But here, we’ve really started to gain insight into the therapeutic angle of how drugs can target these important viral assemblies.”
The researchers are planning additional work on the experimental drugs, focusing on the compound known as 4d, which, based on both pre-clinical testing and the new structural insight, shows more promise against HIV than other compounds. They also want to better understand what happens to the structure of the intasome in cases where it develops resistance to INSTIs. This could help them design more efficient drugs in the future, says Lyumkis.
The findings were published in Science.
Related topics
Drug Development, Drug Targets, Imaging, Structural Biology, Targets, Therapeutics
Related conditions
HIV
Related organisations
Salk Institute
Related people
Dario Passos, Dmitry Lyumkis