Binding site of drug compound and tumour suppressor protein revealed
Posted: 21 April 2020 | Victoria Rees (Drug Target Review) | No comments yet
Using cyro-electron microscopy, researchers have imaged the binding site between a molecule and the tumour suppressor protein PP2A, enabling optimisation of the drug compound.

A team of American scientists has identified the binding site where drug compounds could activate a key braking mechanism against the growth of many types of cancer. According to the researchers, the discovery marks a critical step toward developing a potential new class of anti-cancer drugs that enhance the activity of a prevalent family of tumour suppressor proteins.
The study was led by the University of Michigan Rogel Cancer Center and Case Comprehensive Cancer Center.
Scientists have previously found that certain molecules are capable of increasing the activity of the tumour suppressor protein PP2A, killing cancer cells and shrinking tumours in cell lines and animal models. However, without information about the physical site where the molecules interact with the protein, trying to optimise their properties to turn them into drugs requires endless trial and error.
“We used cryo-electron microscopy to obtain three-dimensional (3D) images of our tool-molecule, DT-061, bound to PP2A,” explained study co-senior author Dr Derek Taylor, an associate professor at Case Western Reserve University and member of the Case Comprehensive Cancer Center. “This allowed us to see for the first time precisely how different parts of the protein were brought together and stabilised by the compound. We can now use that information to start developing compounds that could achieve the desired profile, specificity and potency to potentially translate to the clinic.”
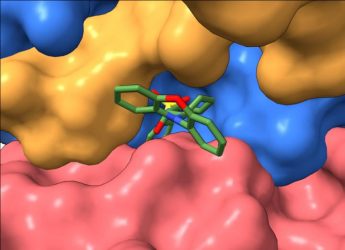

A view of the tool-compound docked to PP2A [credit: Derek Taylor Lab].
The researchers propose calling this class of molecules SMAPs (small molecule activators of PP2A).
In recent years, much research has been invested in the development of kinase inhibitors, small molecule compounds that seek the protein kinases whose dysfunction is involved in the explosive growth and proliferation of cancer cells – cancer’s ‘off-switch’.
The new research instead attacks cancer from the opposite side of the equation, turning on cancer’s off-switch by stabilising protein phosphatases whose malfunction removes a key brake on cancer growth.
The researchers speculate how a combination of both approaches simultaneously might offer an even more powerful strategy.
“The binding pocket we identified provides a launch pad for optimising the next generation of SMAPs toward use in the clinic – in cancer and potentially other diseases,” said research scientist Dr Wei Huang, of the Taylor lab.
The findings appear in Cell.
Related topics
Drug Development, Drug Targets, Kinases, Microscopy, Oncology, Research & Development, Small molecule, Target Molecule
Related conditions
Cancer
Related organisations
Case Comprehensive Cancer Center, University of Michigan Rogel Cancer Center
Related people
Dr Derek Taylor, Dr Wei Huang