Proteomic map of human lung adenocarcinoma revealed
Posted: 13 July 2020 | Hannah Balfour (Drug Target Review) | No comments yet
The proteomic map based on data from 103 patients reveals novel prognostic biomarkers and potential drug targets for lung adenocarcinoma.
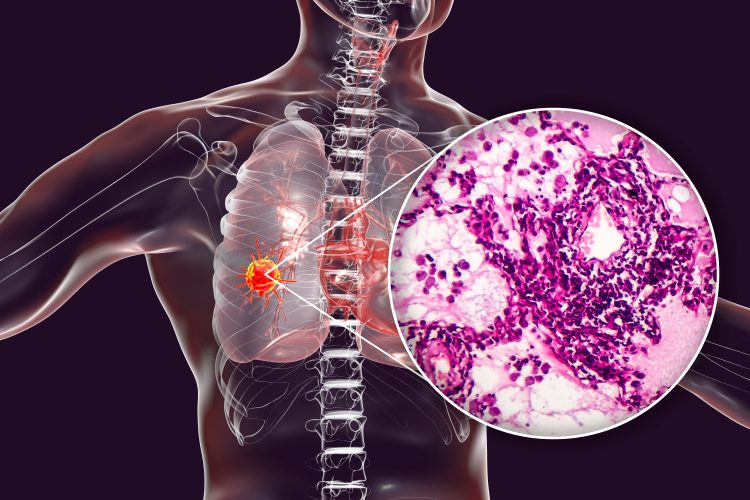

Chinese scientists have revealed a comprehensive proteomic analysis based on 103 patients’ lung adenocarcinoma (LUAD). The research reveals LUAD-related molecular characteristics, as well as potential prognostic biomarkers and drug targets.
The Chinese research institutes who collaborated on the project said their proteomic study of LUAD could fill the gap between genomic abnormality and oncogenic protein function, which has so far prevented the development of targeted therapeutics for the cancer.
The team performed integrated proteomic and phosphoproteomic analyses of tumour tissues and their paired noncancerous adjacent tissues from 103 LUAD patients. A total of 11,119 proteins and 22,564 phosphorylation sites were identified. Combining these proteomics data with clinical information and genomics data, a comprehensive molecular landscape of LUAD was obtained.
The researchers discovered that epithelial-mesenchymal transition, as well as inflammatory and oncogenic signalling pathways, likely led to poor prognosis in stage I LUAD. They also established connections between genetic alterations and proteomic characteristics in patients with mutation of epidermal growth factor receptor (EGFR) or TP53, two predominant driver genes in Chinese LUAD patients.
TTF-1 and Napsin-A, two histological biomarkers, were expresses at elevated levels in LUAD tissues, as were proteins in spliceosomes of patients with EGFR mutation. In patients with TP53 mutation, several oncogenic pathways – including DNA replication and mismatch repair – were noticeably altered.
The team also classified each of the patients into three proteomic subtypes (S-I, S-II, S-III), each containing distinct molecular and clinical features, as well as differing prognoses: S-I was associated with good prognosis, where S-III had poor prognosis.
The phosphoproteomic analysis further revealed activated signalling pathways and kinases in different proteomic subtypes. The integrative analysis of proteomics data and clinical data revealed 27 potential plasma biomarkers and several drug targets for LUAD. One of the potential biomarkers, HSP 90β, was further validated in an independent cohort.
The team concluded that the new proteomic dataset serves as a valuable resource for expediting the translation of research into more precise diagnostics and therapeutic treatments in clinic.
The study was published in Cell.
The research was performed by a team of Chinese scientists from the State Key Laboratory of Drug Research, Shanghai Institute of Materia Medica under the Chinese Academy of Sciences, the State Key Laboratory of Proteomics, National Center for Protein Sciences (Beijing), National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, and Shanghai Jiao Tong University.
Related topics
Biomarkers, Disease Research, Drug Targets, Oncology, Protein, Protein Expression, Proteomics
Related conditions
Lung adenocarcinoma (LAUD), Lung cancer