Study reveals 10 SARS-CoV-2 epitopes that could be targeted by antibodies
Posted: 30 September 2020 | Victoria Rees (Drug Target Review) | No comments yet
An analysis of SARS-CoV-2 has allowed researchers to identify epitopes recognised by a large fraction of COVID-19 patients, with 10 that could be targeted by antibodies.
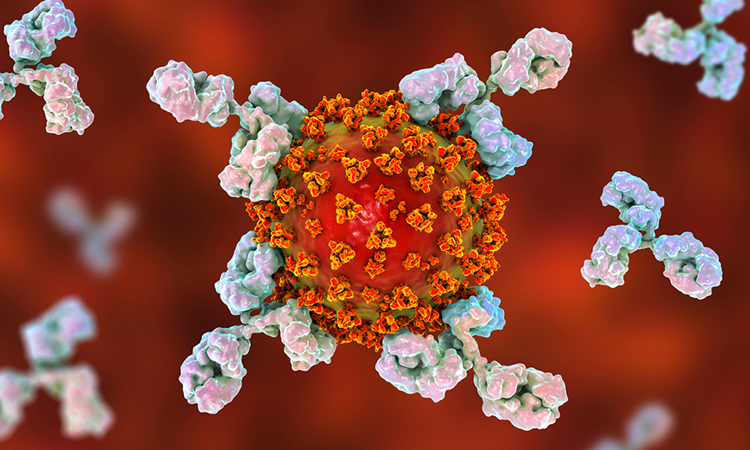

Studying SARS-CoV-2, researchers have identified epitopes recognised by a large fraction of COVID-19 patients, epitopes cross-reactive with antibodies developed in response to other human coronaviruses and 10 epitopes likely recognised by neutralising antibodies.
The study was conducted by a team from Harvard University and John Hopkins University, among other institutions, all in the US.
The team used a technology called VirScan – a tool members of the same group developed previously – to study antibody responses in a large cohort of patients infected with SARS-CoV-2 and control individuals. They used this VirScan data to design a tool for rapid SARS-CoV-2 antibody detection. According to the scientists, the clinical course of COVID-19 is notable for its extreme variability.
Ellen Shrock and colleagues explored the antibody response to SARS-CoV-2 and other human coronaviruses in more than 200 COVID-19 patients and nearly 200 pre-COVID-19 era controls. They found that blood serum from COVID-19 patients exhibited much more SARS-CoV-2 reactivity compared to pre-COVID-19 era controls, though some cross-reactivity toward SARS-CoV-2 peptides was observed in the pre-COVID-19 era samples. They researchers say that this was expected, as nearly everyone has been exposed to human coronaviruses.
However, the team found that only COVID-19 patients’ antibodies primarily recognised peptides derived from the Spike (S) protein and Nucleoprotein (N) of the virus. Those members of the SARS-CoV-2-infected group who had been hospitalised for their infection exhibited stronger and broader antibody responses to S and N peptides, the authors report. In the same group, antibody responses to nearly all viruses, except SARS-CoV-2, were weaker; also, they found that males exhibited greater SARS-CoV-2 antibody responses than females. This latter finding, consistent with reported differences in disease outcomes for males and females, may indicate that males in this group are less able to control the virus soon after infection.
A machine learning model trained on the VirScan results accurately classified patients infected with SARS-CoV-2. The researchers report that drawing insights from this model about peptides highly predictive of SARS-CoV-2 guided the design of simple, rapid diagnostic for COVID-19, which predicted SARS-CoV-2 exposure with 90 percent sensitivity and 95 percent specificity.
“Altogether these data help explain why many serological assays for SARS-CoV-2 produce false positives and should be taken as a cautionary note for those trying to develop such assays,” the authors write.
The study was published in Science.
Related topics
Antibodies, Antibody Discovery, Disease Research, Drug Targets, Peptide Therapeutics, Research & Development
Related conditions
Covid-19
Related organisations
Harvard University, John Hopkins University