Raman spectroscopy platform developed to characterise IDPs
Posted: 29 April 2021 | Victoria Rees (Drug Target Review) | No comments yet
Optical tweezers-coupled Raman spectroscopy has been used to reveal the structure of alpha-synuclein, an IDP linked to Parkinson’s disease.
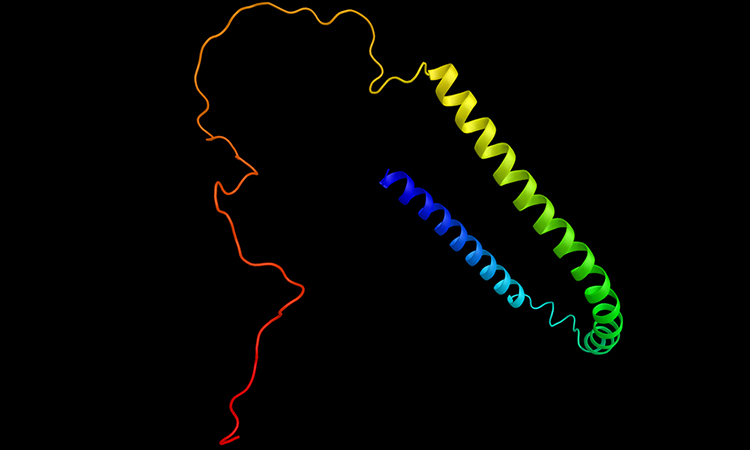

Alpha-synuclein
A research team at the Hong Kong University of Science and Technology (HKUST), has developed optical tweezers-coupled Raman spectroscopy that can directly probe the structural features of alpha-synuclein, an intrinsically disordered protein (IDP) closely linked to Parkinson’s disease, at the physiological concentration by focusing on individual protein molecules.
According to the researchers, IDPs play an important role in biological processes and many of them are associated with incurable neurodegenerative diseases. As a typical IDP, alpha-synuclein lacks a stable three-dimensional (3D) architecture known as secondary structures. It spontaneously undergoes conversions from one secondary structure to another, which could eventually result in the build-up of protein aggregates involved in Parkinson’s disease pathology. However, the transient species during the conversion possess various structures and exist in low population among a dynamic equilibrium mixture. Therefore, their structural features are usually buried under the detection results from traditional measurement techniques, which average the signals detected from large sample quantities and long detection time.
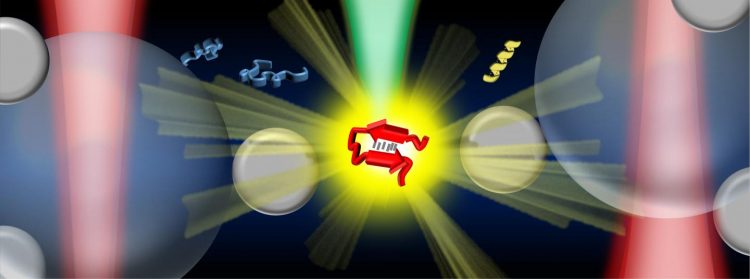

An illustration showing the optical tweezers-controlled hotspot for the protein structural characterisation by surface-enhanced Raman spectroscopy [credit: Vince St Dollente Mesias, Jinqing Huang/HKUST].
In the study, the team integrated optical tweezers and surface-enhanced Raman spectroscopy (SERS) in a novel platform to generate tuneable and reproducible SERS enhancements with single-molecule level sensitivity in aqueous environments, in order to characterise these IDPs while maintaining their intrinsic heterogeneity with great biological significance. Specifically, a hotspot can be visualised and controlled by optical tweezers to allow proteins to go through in a microfluidic flow chamber, which makes it convenient to adjust the measurement parameters in the real time for the in situ spectroscopic characterisations.
The technique directly identified the structural features of the transient species of alpha-synuclein among its predominant monomers at physiological concentration of 1μM by reducing the ensemble averaging in quantity and in time, providing profound insight to understand the initiation of amyloid protein aggregation. Hence, the researchers say this SERS platform has great potential to reveal the structural information of IDPs in the dynamic, heterogeneous and complex biological systems.
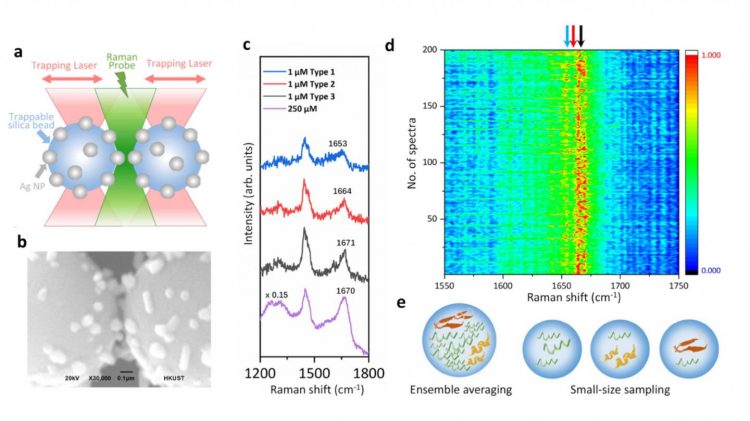

Illustration of the optical tweezers-coupled Raman spectroscopy platform and SERS spectra of alpha-synuclein at physiological concentration showing the structural variations arisen from its transient species [credit: Vince St Dollente Mesias, Jinqing Huang/HKUST].
“Our strategy enables the precise control of the hotspot between two trapped micrometre-size silver nanoparticle-coated silica beads to improve the SERS efficiency and reproducibility in aqueous detections. Except for the tuneable SERS enhancement, the integrated optical tweezers also offer sub-nanometre spatial resolution and sub-piconewton force sensitivity to monitor light-matter interactions in the plasmonic hotspot for extra physical insight. More importantly, our method opens a new door to characterise the transient species of IDPs in dilute solutions, which remains a significant challenge in the biophysics community. Ultimately, it will be exciting to fully exploit the precise force manipulation of the integrated optical tweezers to unfold a single protein inside the controllable hotspot and resolve its structural dynamics from the endogenous molecular vibrations by the integrated Raman spectroscopy,” said Professor Huang Jinqing, who led the research.
The study was published in Nature Communications.
Related topics
Imaging, Label-Free, Spectroscopy
Related conditions
Parkinson's disease
Related organisations
Hong Kong University of Science and Technology (HKUST)
Related people
Professor Huang Jinqing