Method for high-throughput RNA sequencing in single cells developed
Posted: 2 June 2021 | Victoria Rees (Drug Target Review) | No comments yet
A new method named scifi-RNA-seq marks the RNA of cells with specific barcodes, enabling the sequencing of many cells at the same time.
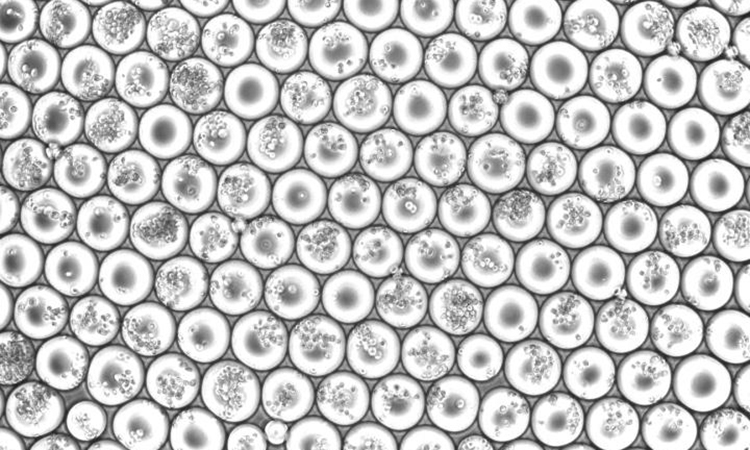

Emulsion droplets from a scifi-RNA-seq experiment loaded with multiples of cells [credit: Paul Datlinger, CeMM].
Scientists from the CeMM Research Center for Molecular Medicine of the Austrian Academy of Sciences have developed a new method for sequencing huge numbers of RNA in single cells in an efficient manner.
According to the team, single-cell RNA sequencing reaches its limits in very large projects, as it is time-consuming and very expensive.
To address these challenges, the researchers developed their new method, called single-cell combinatorial fluidic indexing (scifi-RNA-seq). This marks the RNA of many cells with specific barcodes, before the cells are loaded on a microfluidic chip and their RNA is prepared for single-cell sequencing. These additional barcodes overcome an important problem with existing methods for single-cell sequencing, where single cells are packed into tiny emulsion droplets and assigned cell-specific barcodes. When several cells land in the same droplet, they receive the same barcode and can no longer be distinguished. Therefore, the single cell suspension is loaded onto the microfluidic chip at low concentrations, which means that most of the emulsion droplets remain empty and the reagents are used very inefficiently.
In the scifi-RNA-seq method, the additional barcode means that the emulsion droplets can be loaded with many cells at the same time, while it is still possible to analyse individual cells.
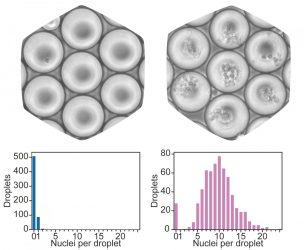

Microfluidic chips for single-cell sequencing can be overloaded 100-fold [credit: Paul Datlinger, CeMM].
“On the popular 10x Genomics system, we use this method to measure 15 times more individual cells. The additional barcode also allows the user to mark and combine thousands of samples in advance and to process those samples together in a single microfluidic analysis. As part of our study, we performed a CRISPR screen with single-cell RNA sequencing readout in human T cells. In the future, our method may, among other things, help to improve immunotherapies for the treatment of cancer,” said study author Paul Datlinger.
“scifi-RNA-seq enables efficient RNA sequencing for millions of individual cells, which facilitates the characterisation of complex tissues, organs and entire organisms. Moreover, in biomedicine it is often useful to analyse many single cells, for example to discover rare stem cells in tumours or cancer cells in the blood. Finally, scifi-RNA-seq will contribute to the trend that drug screens and CRISPR screens are increasingly combined with high-resolution single-cell sequencing readouts,” said project leader Professor Christoph Bock.
The study is published in Nature Methods.
Related topics
Next-Generation Sequencing (NGS), RNAs, Sequencing
Related organisations
CeMM Research Center for Molecular Medicine of the Austrian Academy of Sciences
Related people
Paul Datlinger, Professor Christoph Bock